Chapter 1. In the Beginning
We would like to begin with a great first sentence, like “It was the best of times, it was the worst of times…” but honestly, it’s just the best of times—data science is flourishing! As it continues to mature, it has begun to splinter into niche topics, as many disciplines do over time. This maturity is the result of a long journey that began in the early days of scientific computing. It’s our belief that knowing some of Python’s and R’s origin stories will help you to appreciate how they differ in today’s environment and, thus, how to get the most out of them.
We’re not going to pretend to be science historians, that niche group of academics who trace the circumstances of great discoveries and personalities. What we can do is offer a highlight reel of where Python and R come from and how that led us to our current situation.
The Origins of R
Whenever I think about R, I’m reminded of FUBU, a streetwear company founded back in the 1990s. The name is an acronym that I immediately fell in love with: For Us, By Us. FUBU meant community; it meant understanding the needs and desires of your people and making sure you served them well. R is FUBU.1 By the end of this chapter, I’m sure you’ll feel the same way. Once we acknowledge that R is FUBU, it starts to make a lot more sense.
We can trace the origins of R right back to the now legendary Bell Laboratories in New Jersey. In 1976, development of the statistical programming language S was being spearheaded by John Chambers. A year later, Chambers published Computational Methods for Data Analysis (John Wiley & Sons) and his colleague John Tukey, also at Bell Laboratories, published Exploratory Data Analysis (Addison-Wesley). In 1983, Chambers et al. published Graphical Methods for Data Analysis (CRC Press). These books provided the framework to develop a computational system that would allow a statistician to not only explore, understand, and analyze their data, but also to communicate their results. We’re talking about an all-star FUBU lineup here! Coauthors of Chambers included both Tukey’s cousin Paul A. Tukey and William Cleveland. Cleveland’s empirical experiments on perception, summarized in two insightful books, continue to inform the broader field of data visualization to this day. Among their many contributions to scientific computing and statistics, Tukey developed novel visualizations, like the oft misunderstood box and whiskers plot, and Cleveland developed the LOESS (Locally Weighted Scatterplot Smoothing) method for nonparametric smoothing.
We begin with S because it laid the foundations for what would eventually become R. The nuggets of information in the previous paragraph tell us quite a bit about S’s—and R’s—foundations. First, statisticians are very literal people (S, get it?). This is a pretty helpful trait. Second, statisticians wanted a FUBU programming language specializing in data analysis. They weren’t interested in making a generalist programming language or an operating system. Third, these early books on computational statistics and visualization are, simply put, stunning examples of pedagogical beauty and precise exposition.2 They have aged surprisingly well, despite the obviously dated technology. I’d argue that these books planted the seed for how statisticians, and the R community in particular, approached technical communication in an open, clear, and inclusive manner. This, I believe, is an outstanding and distinctive hallmark of the R community that has deep roots. Fourth, the early emphasis on graphical methods tells us that S was already concerned with flexible and efficient data visualizations, necessary for both understanding data and communicating results. So S was about getting the most important things done as easily as possible, and in a true FUBU way.
The original distribution of S ran on Unix and was available for free. Eventually, S became licensed under an implementation titled S-PLUS. This prompted another open source and free implementation of S by Ross Ihaka and Robert Gentleman at the University of Auckland in 1991. They called this implementation R, for the initials of their first names, as a play on the name S, and in keeping with the tradition of naming programming languages using a single letter. The first official stable beta release of R v1.0.0 was available on February 29, 2000. In the intervening years, two important developments occurred. First, CRAN, the Comprehensive R Archive Network, was established to host and archive R packages on mirrored servers. Second, the R Core Team was also established. This group of volunteers (which currently consists of 20 members) implements base R, including documentation, builds, tests, and releases, plus the infrastructure that makes it all possible. Notably, some of the original members are still involved, including John Chambers, Ross Ihaka, and Robert Gentleman.
A lot has happened since R v1.0.0 in 2000, but the story so far should already give you an idea of R’s unique background as a FUBU statistical computing tool. Before we continue with R’s story, let’s take a look at Python.
The Origins of Python
In 1991, as Ross Ihaka and Robert Gentleman began working on what would become R, Guido van Rossum, a Dutch programmer, released Python. Python’s core vision is really that of one person who set out to address common computing problems at the time. Indeed, van Rossum was lovingly referred to as the benevolent dictator for life (BDFL) for years, a title he gave up when he stepped down from Python’s Steering Council in 2018.
We saw how S arose out of the need for statisticians to perform data analysis and how R arose from the need for an open source implementation, so what problem was addressed by Python? Well, it wasn’t data analysis—that came much later. When Python came on the scene, C and C++, two low-level programming languages, were popular. Python slowly emerged as an interpreted, high-level alternative, in particular after Python v2 was released in 2000 (the same year R v1.0.0 was released). Python was written with the explicit purpose to be, first and foremost, an easy to use and learn, widely adopted programming language with simple syntax. And it has succeeded in this role very well!
This is why you’ll notice that, in contrast to R, Python is everywhere and is incredibly versatile. You’ll see it in web development, gaming, system administration, desktop applications, data science, and so on. To be sure, R is capable of much more than data analysis, but remember, R is FUBU. If R is FUBU, Python is a Swiss Army knife. It’s everywhere and everyone has one, but even though it has many tools, most people just use a single tool on a regular basis. Although data scientists using Python work in a large and varied landscape, they tend to find their niche and specialize in the packages and workflows required for their work instead of exploiting all facets of this generalist language.
Python’s widespread popularity within data science is not entirely due to its data science capabilities. I would posit that Python entered data science by partly riding on the back of existing uses as a general-purpose language. After all, getting your foot in the door is halfway inside. Analysts and data scientists would have had an easier time sharing and implementing scripts with colleagues involved in system administration and web development because they already knew how to work with Python scripts. This played an important role in Python’s widespread adoption. Python was well suited to take advantage of high-performance computing and efficiently implement deep learning algorithms. R was, and perhaps still is, a niche and somewhat foreign language that the wider computing world didn’t really get.
Although Python v2 was released in 2000, a widely adopted package for handling array data didn’t take root until 2005, with the release of NumPy. At this time, SciPy, a package that has, since 2001, provided fundamental algorithms for data science (think optimization, integration, differential equations, etc.), began relying on NumPy data structures. SciPy also provides specialized data structures such as k-dimensional trees.
Once the issue of a standard package for core data structures and algorithms was settled, Python began its ascent into widespread use in scientific computing. The low-level NumPy and SciPy packages laid the foundation for high-level packages like pandas in 2009, providing tools for data manipulation and data structures like data frames. This is sometimes termed the PyData stack, and it’s when the ball really got rolling.
The Language War Begins
The early 2000s set the stage for what some would later refer to as the language wars. As the PyData stack started to take shape, milestones in both Python and R began to heat things up. Four stand out in particular.
First, in 2002, BioConductor was established as a new R package repository and framework for handling the burgeoning (read absolute explosion of) biological data in its myriad forms. Until this point, bioinformaticians relied on tools like MATLAB and Perl (along with classic command-line tools and some manual web-interface tools). MATLAB is still favored in specific disciplines, like neuroscience. However, Perl has been mostly superseded by BioConductor. BioConductor’s impact on bioinformatics is hard to overstate. Not only did it provide a repository of packages for dealing with remote genetic sequence databases, expression data, microarrays, and so on, it also provided new data structures to handle genetic sequences. BioConductor continues to expand and is deeply embedded within the bioinformatics community.
Second, in 2006 the IPython package was released. This was a groundbreaking way to work on Python in an interactive notebook environment. Following various grants beginning in 2012, IPython eventually matured into the Jupyter Project in 2014, which now encompasses the JupyterLab IDE. Users often forget that Jupyter is short for “Julia, Python, and R” because it’s very Python-centric. Notebooks have become a dominant way of doing data science in Python, and in 2018 Google released Google Colab, a free online notebook tool. We’ll dig into this in Chapter 3.
Third, in 2007, Hadley Wickham published his PhD thesis, which consisted of two R packages that would fundamentally change the R landscape. The first, reshape, laid the foundations for what would later become formalized as the Tidyverse (more on this later). Although reshape has long since been retired, it was the first glimpse into understanding how data structure influences how we think about and work with our data.3 The second, ggplot2, is an implementation of the seminal book by Leland Wilkinson et al., The Grammar of Graphics (Springer), and provided intuitive, high-level plotting that greatly simplified previously existing tools in R (more on this in Chapter 5).
Finally, Python v3 was released in 2008. For years the question persisted as to which version of Python to use, v2 or v3. That’s because Python v3 is backward-incompatible.4 Luckily, this has been resolved for you since Python v2 was retired in 2020. Surprisingly, you can still buy a new MacBook Pro after that date with Python 2 preinstalled because legacy scripts still rely on it. So Python 2 lives on still.
The Battle for Data Science Dominance
By this point both Python and R had capable tools for a wide variety of data science applications. As the so-called “language wars” continued, other key developments saw each language find its niche.
Both Python and R were wrapped up in specific builds. For Python this was the Anaconda distribution, which is still in popular use (see Chapter 3). For R, Revolution Analytics, a data science software developer, produced Revolution R Open. Although their R build was never widely adopted by the community, the company was acquired by Microsoft, signaling strong corporate support of the R language.
In 2011, the Python community foresaw the boom in machine learning with the release of the scikit-learn package. In 2016, this was followed by the release of both TensorFlow and Keras for deep learning, also with a healthy dose of corporate support. This also highlights Python’s strength as a high-level interpreter sitting on top of high-performance platforms. For example, you’ll find Amazon Web Services (AWS) Lambda for massive highly concurrent programming, Numba for high-performance computing, and the aforementioned TensorFlow for highly optimized C++. With its widespread adoption outside of data science, it’s no surprise that Python gained a reputation for deploying models in a way that R could not.
2011 also saw the release of RStudio IDE by the eponymous company, and over the next few years the R community began to converge on this tool. At this point, to use R is, in many regards, to use RStudio. The influence RStudio has on promoting R as a programming language suitable for a wide variety of data-centric uses is also important to note.
While all of this was happening, a growing segment of the R community began to move toward a suite of packages, many of which were authored or spearheaded by Hadley Wickham, that began to reframe and simplify typical data workflows. Much of what these packages did was to standardize R function syntax, as well as input and output data storage structures. Eventually the suite of packages began to be referred to colloquially as the “Hadleyverse.” In a keynote speech at the useR! 2016 conference at Stanford University, Wickham did away with this, igniting digital flames to burn up his name and coining the term “Tidyverse.” Since Wickham joined RStudio, the company has been actively developing and promoting the Tidyverse ecosystem, which has arguably become the dominant dialect in R. We’ll explore this in more depth in Chapter 2.
We can imagine that R contains at least two “paradigms,” or “dialects.” They can be mixed, but each has its own distinct flavor. Base R is what most R has been and, probably, still is. Tidyverse reimagines base R in a broad, all-encompassing universe of packages and functions that play well together, often relying on piping,5 and has a preference for data frames.6 I would argue that BioConductor provides yet another dialect, which is focused on a specific discipline, bioinformatics. You’ll no doubt find that some large packages may contain enough idiosyncrasies that you may consider them a dialect in their own right, but let’s not go down that rabbit hole. R is now at the threshold where some users know (or are taught) only the Tidyverse way of doing things. The distinction between base and Tidyverse R may seem trivial, but I have seen many new R learners struggle to make sense of why the Tidyverse exists. This is partly because years of base R code is still in active use and can’t be ignored. Although Tidyverse advocates argue that these packages make life much easier for the beginner, competing dialects can cause unnecessary confusion.
We can also imagine that Python contains distinct dialects. The vanilla installation of Python is the bare-bones installation, and operates differently from an environment that has imported the PyData stack. For the most part, data scientists operate within the PyData stack, so there’s less confusion between dialects.
A Convergence on Cooperation and Community-Building
For a time, it seemed that the prevailing attitude in the language wars was an us versus them mentality. A look of disdain glancing at a person’s computer screen. It seemed like either Python or R would eventually disappear from the data science landscape. Hello monoculture! Some data scientists are still rooting for this, but we’re guessing you’re not one of them. And there was also a time when it seemed like Python and R were trying to mimic each other, just porting workflows so that language didn’t matter. Luckily those endeavors have not come to fruition. Both Python and R have unique strengths; trying to imitate each other seems to miss that point.
Today many data scientists in the Python and R communities recognize that both languages are outstanding, useful, and complementary. To return to a key point in the preface, the data science community has converged onto a point of cooperation and community-building—to the benefit of everyone involved.
We’re ready for a new community of bilingual data scientists. The challenge is that many users of one language don’t quite know how they are complementary or when to use which language. There have been a few solutions over the years, but we’ll get into that in Part IV.
Final Thoughts
At this point you should have a good idea of where we are in 2021 and how we got here. In the next part we’ll introduce each group of users to a new language.
One last note: Python users refer to themselves as Pythonistias, which is a really cool name! There’s no real equivalent in R, and they also don’t get a really cool animal, but that’s life when you’re a single-letter language. R users are typically called…wait for it…useRs! (Exclamation optional.) Indeed, the official annual conference is called useR! (exclamation obligatory), and the publisher Springer has an ongoing and very excellent series of books of the same name. We’ll use these names from now own.
Figure 1-1 provides a summary of some of the major events that we’ve highlighted in this chapter, plus some other milestones of interest.
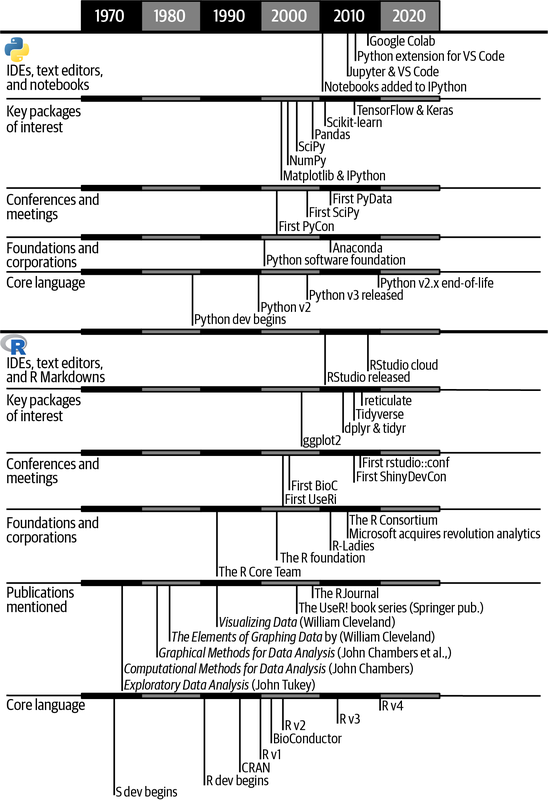
Figure 1-1. A timeline of Python and R data science milestones
1 Well, OK, more like For Statisticians, By Statisticians, but FSBS doesn’t have the same ring to it.
2 With the possible exception of Computational Methods for Data Analysis, which I admit to not having read.
3 I would argue that we can trace this relationship back to the early days of R, as evidenced by formula notation and various built-in datasets. Nonetheless, a consistent and intuitive framework was lacking.
4 This has even moved some prominent developers to voice an aversion to the eventual development of Python 4.0. How Python develops will be exciting to watch!
5 That is, using an output of one function as the input of another function.
6 Python users might not be familiar with the term base. This means only the built-in functionality of the language without any additional package installations. Base R itself is well equipped for data analysis. In Python, a data scientist would import the PyData stack by default.
Get Python and R for the Modern Data Scientist now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

