Chapter 4. Working with File-Based and Feed-Based Data in Python
In Chapter 3, we focused on the many characteristics that contribute to data quality—from the completeness, consistency, and clarity of data integrity to the reliability, validity, and representativeness of data fit. We discussed the need to both “clean” and standardize data, as well as the need to augment it by combining it with other datasets. But how do we actually accomplish these things in practice?
Obviously, it’s impossible to begin assessing the quality of a dataset without first reviewing its contents—but this is sometimes easier said than done. For decades, data wrangling was a highly specialized pursuit, leading companies and organizations to create a whole range of distinct (and sometimes proprietary) digital data formats designed to meet their particular needs. Often, these formats came with their own file extensions—some of which you may have seen: xls, csv, dbf, and spss are all file formats typically associated with “data” files.1 While their specific structures and details vary, all of these formats are what I would describe as file-based—that is, they contain (more or less) historical data in static files that can be downloaded from a database, emailed by a colleague, or accessed via file-sharing sites. Most significantly, a file-based dataset will, for the most part, contain the same information whether you open it today or a week, a month, or a year from now.
Today, these file-based formats stand in contrast to the data formats and interfaces that have emerged alongside real-time web services over the past 20 years. Web-based data today is available for everything from news to weather monitoring to social media sites, and these feed-style data sources have their own unique formats and structures. Extensions like xml, json, and rss indicate this type of real-time data, which often needs to be accessed via specialized application programming interfaces, or APIs. Unlike file-based formats, accessing the same web-based data location or “endpoint” via an API will always show you the most recent data available—and that data may change in days, hours, or even seconds.
These aren’t perfect distinctions, of course. There are many organizations (especially government departments) that provide file-based data for download—but then overwrite those files with new ones that have the same name whenever the source data is updated. At the same time, feed-style data formats can be downloaded and saved for future reference—but their source location online will not generally provide access to older versions. Despite these sometimes unconventional uses for each class of data format, however, in most cases you can use the high-level differences between file-based and feed-based data formats to help you choose the most appropriate sources for a given data wrangling project.
How do you know if you want file-based or feed-based data? In many cases, you won’t have a choice. Social media companies, for example, provide ready access to their data feeds through their APIs but don’t generally provide retrospective data. Other types of data—especially data that is itself synthesized from other sources or heavily reviewed before release—are much more likely to be made available in file-based formats. If you do have a choice between file-based and feed-based formats, then which you choose will really depend on the nature of your data wrangling question: if it hinges on having the most recent data available, then a feed-style format will probably be preferable. But if you’re concerned about trends, file-based data, which is more likely to contain information collected over time, will probably be your best bet. That said, even when both formats are available, there’s no guarantee they contain the same fields, which once again might make your decision for you.
Over the course of this chapter, we’ll work through hands-on examples of wrangling data from several of the most common file-based and feed-based data formats, with the goal of making them easier to review, clean, augment, and analyze. We’ll also take a look at some of the tougher-to-wrangle data formats that you might need to work with strictly out of necessity. Throughout these processes, we’ll rely heavily on the excellent variety of libraries that the Python community has developed for these purposes, including specialty libraries and programs for processing everything from spreadsheets to images. By the time we finish, you’ll have the skills and sample scripts you need to tackle a huge variety of data wrangling projects, paving the way forward for your next data wrangling project!
Structured Versus Unstructured Data
Before we dive into writing code and wrangling data, I want to briefly discuss one other key attribute of data sources that can impact the direction (and speed) of your data wrangling projects—working with structured versus unstructured data.
The goal of most data wrangling projects is to generate insight and, often, to use data to make better decisions. But decisions are time sensitive, so our work with data also requires balancing trade-offs: instead of waiting for the “perfect” dataset, we may combine two or three not-so-perfect ones in order to build a valid approximation of the phenomenon we’re investigating, or we may look for datasets that share common identifiers (for example, zip codes), even if that means we need to later derive the particular dimensional structure (like neighborhood) that truly interests us. As long as we can gain these efficiencies without sacrificing too much in terms of data quality, improving the timeliness of our data work can also increase its impact.
One of the simplest ways to make our data wrangling more efficient is to seek out data formats that are easy for Python and other computational tools to access and understand. Although advances in computer vision, natural language processing, and machine learning have made it easier for computers to analyze data regardless of its underlying structure or format, the fact is that structured, machine-readable data remains—unsurprisingly, perhaps—the most straightforward type of data to work with. In truth, while anything from interviews to images to the text of books can be used as a data source, when many of us think of “data,” we often think of structured, numerical data more than anything else.
Structured data is any type of data that has been organized and classified in some way, into some version of records and fields. In file-based formats, these are usually rows and columns; in feed-based formats they are often (essentially) lists of objects or dictionaries.
Unstructured data, by contrast, may consist of a mash-up of different data types, combining text, numbers, and even photographs or illustrations. The contents of a magazine or a novel, or the waveforms of a song, for example, would typically be considered unstructured data.
If right now you’re thinking, “Hang on, novels have structure! What about chapters?” then congratulations: you’re already thinking like a data wrangler. We can create data about almost anything by collecting information about the world and applying structure to it.4 And in truth, this is how all data is created: the datasets that we access via files and feeds are all the product of someone’s decisions about how to collect and organize information. In other words, there is always more than one way to organize information, but the structure chosen influences how it can be analyzed. This is why it’s a little ridiculous to suggest that data can somehow be “objective”; after all, it’s the product of (inherently subjective) human choices.
For example, try conducting this mini-experiment: think about how you organize some collection of yours (it could be a collection of music, books, games, or varieties of tea—you name it). Now ask a friend how they organize their own collection of that item. Do you do it the same way? Which is “better”? Now ask someone else, and maybe even a third person. While you may find similarities among the systems that you and your friends use for organizing your music collections, for example, I would be very surprised if you find that any two of you do it precisely the same way. In fact, you’ll probably find that everyone does it a little bit differently but also feels passionately that their way is “best.” And it is! For them.
If this is reminding you of our discussion in “How? And for Whom?”, that’s no coincidence, because the result of your data wrangling question and efforts will eventually be—you guessed it!—another dataset, which will reflect your interests and priorities. It, too, will be structured and organized, which makes working with it in certain ways easier than others. But the takeaway here is not that any given way is right or wrong, just that every choice involves trade-offs. Identifying and acknowledging those trade-offs is a key part of using data honestly and responsibly.
So a key trade-off when using structured data is that it requires depending on someone else’s judgments and priorities in organizing the underlying information. Obviously, this can be a good—or even great!—thing if that data has been structured according to an open, transparent process that involves well-qualified experts. Thoughtfully applied data structures like this can give us early insight into a subject we may otherwise know little to nothing about. On the other hand, there is also the possibility that we will inherit someone else’s biased or poorly designed choices.
Unstructured data, of course, gives us complete freedom to organize information into data structures that suit our needs best. Unsurprisingly, this requires us to take responsibility for engaging a robust data quality process, which may be both complex and time-consuming.
How do we know if a particular dataset is structured or unstructured up front? In this case, file extensions can definitely help us out. Feed-based data formats always have at least some structure to them, even if they contain chunks of “free text,” like social media posts. So if you see the file extensions .json, .xml, .rss, or .atom, the data has at least some type of record-and-field structure, as we’ll explore in “Feed-Based Data—Web-Driven Live Updates”. File-based data that ends in .csv, .tsv, .txt, .xls(x), or .ods tends to follow a table-type, rows-and-columns structure, as we’ll see in the next section. Truly unstructured data, meanwhile, is most likely to come to us as .doc(x) or .pdf.
Now that we’ve got a good handle on the different types of data sources that we’re likely to encounter—and even some sense of how to locate them—let’s get wrangling!
Working with Structured Data
Since the early days of digital computing, the table has been one of the most common ways to structure data. Even today, many of the most common and easy-to-wrangle data formats are little more than tables or collections of tables. In fact, we already worked with one very common table-type data format in Chapter 2: the .csv or comma-separated value format.
File-Based, Table-Type Data—Take It to Delimit
In general, all of the table-type data formats you’ll typically encounter are examples of what are known as delimited files: each data record is on its own line or row, and the boundaries between fields or columns of data values are indicated—or delimited—by a specific text character. Often, an indication of which text character is being used as the delimiter in a file is incorporated into the dataset’s file extension. For example, the .csv file extension stands for comma-separated value, because these files use a comma character (,) as a delimiter; the .tsv file extension stands for tab-separated value, because the data columns are separated by a tab. A list of file extensions commonly associated with delimited data follows:
- .csv
-
Comma-separated value files are among the most common form of table-type structured data files you’ll encounter. Almost any software system that handles tabular data (such as government or corporate data systems, spreadsheet programs, and even specialized commercial data programs) can output data as a .csv, and, as we saw in Chapter 2, there are handy libraries for easily working with this data type in Python.
- .tsv
-
Tab-separated value files have been around for a long time, but the descriptive .tsv extension has only become common relatively recently. While data providers don’t often explain why they choose one delimiter over another, tab-delimited files may be more common for datasets whose values need to include commas, such as postal addresses.
- .txt
-
Structured data files with this extension are often .tsv files in disguise; older data systems often labeled tab-separated data with the .txt extension. As you’ll see in the worked examples that follow, it’s a good idea to open and review any data file you want to wrangle with a basic text program (or a code editor like Atom) before you write any code, since looking at the contents of the file is the only surefire way to know what delimiters you’re working with.
- .xls(x)
-
This is the file extension of spreadsheets produced with Microsoft Excel. Because these files can contain multiple “sheets” in addition to formulas, formatting, and other features that simple delimited files cannot replicate, they need more memory to store the same amount of data. They also have other limitations (like only being able to handle a certain number of rows) that can have implications for your dataset’s integrity.
- .ods
-
Open-document spreadsheet files are the default extension for spreadsheets produced by a number of open source software suites like LibreOffice and OpenOffice and have limitations and features similar to those of .xls(x) files.
Before we dive into how to work with each of these file types in Python, it’s worth spending just a little time thinking about when we might want to work with table-type data and where to find it when we do.
When to work with table-type data
Most of the time, we don’t get much of a choice about the format of our source data. In fact, much of the reason we need to do data wrangling in the first place is because the data we have doesn’t quite meet our needs. That said, it’s still valuable to know what data format you would prefer to be able to work with so that you can use that to inform your initial search for data.
In “Structured Versus Unstructured Data”, we talked about the benefits and limitations of structured data, and we now know that table-type data is one of the oldest and most common forms of machine-readable data. This history means, in part, that over the years many forms of source data have been crammed into tables, even though they may not necessarily be well suited to table-like representations. Still, this format can be especially useful for answering questions about trends and patterns over time. In our Citi Bike exercise from Chapter 2, for example, we examined how many “Customers” versus “Subscribers” had taken Citi Bike rides over the course of a single month. If we wanted to, we could perform the same calculation for every available month of Citi Bike rides in order to understand any patterns in this ratio over time.
Of course, table-type data is generally not a great format for real-time data, or data where not every observation contains the same possible values. These kinds of data are often better suited to the feed-based data formats that we discuss in “Feed-Based Data—Web-Driven Live Updates”.
Where to find table-type data
Since the vast majority of machine-readable data is still in table-type data formats, it is among the easiest data formats to locate. Spreadsheets are common in every discipline, and a large number of government and commercial information systems rely on software that organizes data in this way. Almost any time you request data from an expert or organization, a table-type format is what you are likely to get. This is also true of almost every open-data portal and data-sharing site you’ll find online. As we covered in “Smart Searching for Specific Data Types”, you can even find table-type data (and other specific file formats) via search engines, if you know how to look.
Wrangling Table-Type Data with Python
To help illustrate how simple it is to work with table-type data in Python, we’ll walk through examples of how to read in data from all of the file types mentioned in this section—plus a few others, just for good measure. While in later chapters we’ll look at how to do more with cleaning, transformation, and data quality assessments, our focus for the time being will simply be on accessing the data within each type of data file and interacting with it using Python.
Reading data from CSVs
In case you didn’t follow along in Chapter 2, here’s a refresher on how to read data from a .csv file, using a sample from the Citi Bike dataset (Example 4-1). As always, I’ve included a description of what the program is doing—as well as links to any source files—in the comments at the top of my script. Since we’ve worked with this data format before, for now we’ll just worry about printing out the first few rows of data to see what they look like.
Example 4-1. csv_parsing.py
# a simple example of reading data from a .csv file with Python# using the "csv" library.# the source data was sampled from the Citi Bike system data:# https://drive.google.com/file/d/17b461NhSjf_akFWvjgNXQfqgh9iFxCu_/# which can be found here:# https://s3.amazonaws.com/tripdata/index.html# import the `csv` libraryimportcsv# open the `202009CitibikeTripdataExample.csv` file in read ("r") mode# this file should be in the same folder as our Python script or notebooksource_file=open("202009CitibikeTripdataExample.csv","r")# pass our `source_file` as an ingredient to the `csv` library's# DictReader "recipe".# store the result in a variable called `citibike_reader`citibike_reader=csv.DictReader(source_file)# the DictReader method has added some useful information to our data,# like a `fieldnames` property that lets us access all the values# in the first or "header" row(citibike_reader.fieldnames)# let's just print out the first 5 rowsforiinrange(0,5):(next(citibike_reader))
This is our workhorse library when it comes to dealing with table-type data.
open()is a built-in function that takes a filename and a “mode” as parameters. In this example, the target file (202009CitibikeTripdataExample.csv) should be in the same folder as our Python script or notebook. Values for the “mode” can berfor “read” orwfor “write.”
By printing out the
citibike_reader.fieldnamesvalues, we can see that the exact label for the “User Type” column isusertype.
The
range()function gives us a way to execute some piece of code a specific number of times, starting with the value of the first argument and ending just before the value of the second argument. For example, the code indented below this line will be executed five times, going through theivalues of0,1,2,3, and4. For more on therange()function, see “Adding Iterators: The range Function”.
The output from running this should look something like:
['tripduration', 'starttime', 'StartDate', 'stoptime', 'start station id',
'start station name', 'start station latitude', 'start station longitude', 'end
station id', 'end station name', 'end station latitude', 'end station
longitude', 'bikeid', 'usertype', 'birth year', 'gender']
{'tripduration': '4225', 'starttime': '2020-09-01 00:00:01.0430', 'StartDate':
'2020-09-01', 'stoptime': '2020-09-01 01:10:26.6350', 'start station id':
'3508', 'start station name': 'St Nicholas Ave & Manhattan Ave', 'start station
latitude': '40.809725', 'start station longitude': '-73.953149', 'end station
id': '116', 'end station name': 'W 17 St & 8 Ave', 'end station latitude': '40.
74177603', 'end station longitude': '-74.00149746', 'bikeid': '44317',
'usertype': 'Customer', 'birth year': '1979', 'gender': '1'}
...
{'tripduration': '1193', 'starttime': '2020-09-01 00:00:12.2020', 'StartDate':
'2020-09-01', 'stoptime': '2020-09-01 00:20:05.5470', 'start station id':
'3081', 'start station name': 'Graham Ave & Grand St', 'start station
latitude': '40.711863', 'start station longitude': '-73.944024', 'end station
id': '3048', 'end station name': 'Putnam Ave & Nostrand Ave', 'end station
latitude': '40.68402', 'end station longitude': '-73.94977', 'bikeid': '26396',
'usertype': 'Customer', 'birth year': '1969', 'gender': '0'}
Reading data from TSV and TXT files
Despite its name, the Python csv library is basically a one-stop shop for wrangling table-type data in Python, thanks to the DictReader function’s delimiter option. Unless you tell it differently, DictReader assumes that the comma character (,) is the separator it should look for. Overriding that assumption is easy, however: you can simply specify a different character when you call the function. In Example 4-2, we specify the tab character (\t), but we could easily substitute any delimiter we prefer (or that appears in a particular source file).
Example 4-2. tsv_parsing.py
# a simple example of reading data from a .tsv file with Python, using# the `csv` library. The source data was downloaded as a .tsv file# from Jed Shugerman's Google Sheet on prosecutor politicians:# https://docs.google.com/spreadsheets/d/1E6Z-jZWbrKmit_4lG36oyQ658Ta6Mh25HCOBaz7YVrA# import the `csv` libraryimportcsv# open the `ShugermanProsecutorPoliticians-SupremeCourtJustices.tsv` file# in read ("r") mode.# this file should be in the same folder as our Python script or notebooktsv_source_file=open("ShugermanProsecutorPoliticians-SupremeCourtJustices.tsv","r")# pass our `tsv_source_file` as an ingredient to the csv library's# DictReader "recipe."# store the result in a variable called `politicians_reader`politicians_reader=csv.DictReader(tsv_source_file,delimiter='\t')# the DictReader method has added some useful information to our data,# like a `fieldnames` property that lets us access all the values# in the first or "header" row(politicians_reader.fieldnames)# we'll use the `next()` function to print just the first row of data(next(politicians_reader))
This dataset was listed in Jeremy Singer-Vine’s (@jsvine) “Data Is Plural” newsletter (https://data-is-plural.com).
This should result in output that looks something like:
['', 'Justice', 'Term Start/End', 'Party', 'State', 'Pres Appt', 'Other Offices
Held', 'Relevant Prosecutorial Background']
{'': '40', 'Justice': 'William Strong', 'Term Start/End': '1870-1880', 'Party':
'D/R', 'State': 'PA', 'Pres Appt': 'Grant', 'Other Offices Held': 'US House,
Supr Court of PA, elect comm for elec of 1876', 'Relevant Prosecutorial
Background': 'lawyer'}
Though the .tsv file extension has become relatively common nowadays, many files generated by older databases that are actually tab-separated may reach you with a .txt file extension. Fortunately, as described in the preceding sidebar, this changes nothing about how we handle the file as long as we specify the correct delimiter—as you can see in Example 4-3.
Example 4-3. txt_parsing.py
# a simple example of reading data from a .tsv file with Python, using# the `csv` library. The source data was downloaded as a .tsv file# from Jed Shugerman's Google Sheet on prosecutor politicians:# https://docs.google.com/spreadsheets/d/1E6Z-jZWbrKmit_4lG36oyQ658Ta6Mh25HCOBaz7YVrA# the original .tsv file was renamed with a file extension of .txt# import the `csv` libraryimportcsv# open the `ShugermanProsecutorPoliticians-SupremeCourtJustices.txt` file# in read ("r") mode.# this file should be in the same folder as our Python script or notebooktxt_source_file=open("ShugermanProsecutorPoliticians-SupremeCourtJustices.txt","r")# pass our txt_source_file as an ingredient to the csv library's DictReader# "recipe" and store the result in a variable called `politicians_reader`# add the "delimiter" parameter and specify the tab character, "\t"politicians_reader=csv.DictReader(txt_source_file,delimiter='\t')# the DictReader function has added useful information to our data,# like a label that shows us all the values in the first or "header" row(politicians_reader.fieldnames)# we'll use the `next()` function to print just the first row of data(next(politicians_reader))
As discussed in “Don’t Leave Space!”, whitespace characters have to be escaped when we’re using them in code. Here, we’re using the escaped character for a
tab, which is\t. Another common whitespace character code is\nfornewline(or\rforreturn, depending on your device).
If everything has gone well, the output from this script should look exactly the same as that from Example 4-2.
One question you may be asking yourself at this point is “How do I know what delimiter my file has?” While there are programmatic ways to help detect this, the simple answer is: Look! Anytime you begin working with (or thinking about working with) a new dataset, start by opening it up in the most basic text program your device has to offer (any code editor will also be a reliable choice). Especially if the file is large, using the simplest program possible will let your device devote maximum memory and processing power to actually reading the data—reducing the likelihood that the program will hang or your device will crash (closing other programs and excess browser tabs will help, too)!
Though I’ll talk about some ways to inspect small parts of really large files later on in the book, now is the time to start practicing the skills that are essential to assessing data quality—all of which require reviewing your data and making judgments about it. So while there are ways to “automate away” tasks like identifying the correct delimiter for your data, eyeballing it in a text editor will often be not just faster and more intuitive, but it will help you get more familiar with other important aspects of the data at the same time.
Real-World Data Wrangling: Understanding Unemployment
The underlying dataset that we’ll use to explore some of our trickier table-type data formats is unemployment data about the United States. Why? In one way or another, unemployment affects most of us, and in recent decades the US has experienced some particularly high unemployment rates. Unemployment numbers for the US are released monthly by the Bureau of Labor Statistics (BLS), and while they are often reported by general-interest news sources, they are usually treated as a sort of abstract indicator of how “the economy” is doing. What the numbers really represent is rarely discussed in-depth.
When I first joined the Wall Street Journal in 2007, building an interactive dashboard for exploring monthly economic indicator data—including unemployment—was my first major project. One of the more interesting things I learned in the process is that there isn’t “an” unemployment rate calculated each month, there are several (six, to be exact). The one that usually gets reported by news sources is the so-called “U3” unemployment rate, which the BLS describes as:
Total unemployed, as a percent of the civilian labor force (official unemployment rate).
On its surface, this seems like a straightforward definition of unemployment: of all the people who reasonably could be working, what percentage are not?
Yet the real story is a bit more complex. What does it mean to be “employed” or be counted as part of the “labor force”? A look at different unemployment numbers makes more clear what the “U3” number does not take into account. The “U6” unemployment rate is defined as:
Total unemployed, plus all persons marginally attached to the labor force, plus total employed part time for economic reasons, as a percent of the civilian labor force plus all persons marginally attached to the labor force.
When we read the accompanying note, this longer definition starts to take shape:5
NOTE: Persons marginally attached to the labor force are those who currently are neither working nor looking for work but indicate that they want and are available for a job and have looked for work sometime in the past 12 months. Discouraged workers, a subset of the marginally attached, have given a job-market related reason for not currently looking for work. Persons employed part time for economic reasons are those who want and are available for full-time work but have had to settle for a part-time schedule. Updated population controls are introduced annually with the release of January data.
In other words, if you want a job (and have looked for one in the past year) but haven’t looked for one very recently—or if you have a part-time job but want a full-time job—then you don’t officially count as “unemployed” in the U3 definition. This means that the economic reality of Americans working multiple jobs (who are more likely to be women and have more children)6, and potentially of “gig” workers (recently estimated as up to 30% of the American workforce),7 are not necessarily reflected in the U3 number. Unsurprisingly, the U6 rate is typically several percentage points higher each month than the U3 rate.
To see how these rates compare over time, we can download them from the website of the St. Louis Federal Reserve, which provides thousands of economic datasets for download in a range of formats, including table-type .xls(x) files and, as we’ll see later in Example 4-12, feed-type formats as well.
You can download the data for these exercises from the Federal Reserve Economic Database (FRED) website. It shows the current U6 unemployment rate since the measure was first created in the early 1990s.
To add the U3 rate to this graph, at the top right choose Edit graph → ADD LINE. In the search field, type UNRATE and then select “Unemployment Rate” when it populates below the search bar. Finally, click Add series. Close this side window using the X at the top right, and then select Download, being sure to select the first option, Excel.8 This will be an .xls file, which we’ll handle last because although still widely available, this is a relatively outdated file format (it was replaced by .xlsx as the default format for Microsoft Excel spreadsheets in 2007).
To get the additional file formats we need, just open the file you downloaded with a spreadsheet program like Google Sheets and choose “Save As,” then select .xlsx, then repeat the process choosing .ods. You should now have the following three files, all containing the same information: fredgraph.xlsx, fredgraph.ods, and fredgraph.xls.9
Note
If you opened the original fredgraph.xls file, you probably noticed that it contains more than just the unemployment data; it also contains some header information about where the data came from and the definitions of U3 and U6 unemployment, for example. While doing analysis on the unemployment rates these files contain would require separating this metadata from the table-type data further down, remember that our goal for the moment is simply to convert all of our various files to a .csv format. We’ll tackle the data cleaning process that would involve removing this metadata in Chapter 7.
XLSX, ODS, and All the Rest
For the most part, it’s preferable to avoid processing data saved as .xlsx, .ods, and most other nontext table-type data formats directly, if possible. If you’re just at the stage of exploring datasets, I suggest you review these files simply by opening them with your preferred spreadsheet program and saving them as a .csv or .tsv file format before accessing them in Python. Not only will this make them easier to work with, it will give you a chance to actually look at the contents of your data file and get a sense of what it contains.
Resaving and reviewing .xls(x) and similar data formats as a .csv or equivalent text-based file format will both reduce the file size and give you a better sense of what the “real” data looks like. Because of the formatting options in spreadsheet programs, sometimes what you see onscreen is substantially different from the raw values that are stored in the actual file. For example, values that appear as percentages in a spreadsheet program (e.g., 10%) might actually be decimals (.1). This can lead to problems if you try to base aspects of your Python processing or analysis on what you saw in the spreadsheet as opposed to a text-based data format like .csv.
Still, there will definitely be situations where you need to access .xls(x) and similar file types with Python directly.10 For example, if there’s an .xls dataset you need to wrangle on a regular basis (say, every month), resaving the file manually each time would become unnecessarily time-consuming.
Fortunately, that active Python community we talked about in “Community” has created libraries that can handle an impressive range of data formats with ease. To get a thorough feel for how these libraries work with more complex source data (and data formats), the following code examples read in the specified file format and then create a new .csv file that contains the same data.
To make use of these libraries, however, you’ll first need to install them on your device by running the following commands one by one in a terminal window:11
pip install openpyxl pip install pyexcel-ods pip install xlrd==2.0.1
In the following code examples, we’ll be using the openpyxl library to access (or parse) .xlsx files, the pyexcel-ods library for dealing with .ods files, and the xlrd library for reading from .xls files (for more on finding and selecting Python libraries, see “Where to Look for Libraries”).
To better illustrate the idiosyncrasies of these different file formats, we’re going to do something similar to what we did in Example 4-3: we’ll take sample data that is being provided as an .xls file and create .xlsx and .ods files containing the exact same data by resaving that source file in the other formats using a spreadsheet program. Along the way, I think you’ll start to get a sense of how these nontext formats make the process of data wrangling more (and, I would argue, unnecessarily) complicated.
We’ll start by working through an .xlsx file in \ref (Example 4-4), using a version of the unemployment data downloaded from FRED. This example illustrates one of the first major differences between dealing with text-based table-type data files and nontext formats: because the nontext formats support multiple “sheets,” we needed to include a for loop at the top of our script, within which we put the code for creating our individual output files (one for each sheet).
Example 4-4. xlsx_parsing.py
# an example of reading data from an .xlsx file with Python, using the "openpyxl"# library. First, you'll need to pip install the openpyxl library:# https://pypi.org/project/openpyxl/# the source data can be composed and downloaded from:# https://fred.stlouisfed.org/series/U6RATE# specify the "chapter" you want to import from the "openpyxl" library# in this case, "load_workbook"fromopenpyxlimportload_workbook# import the `csv` library, to create our output fileimportcsv# pass our filename as an ingredient to the `openpyxl` library's# `load_workbook()` "recipe"# store the result in a variable called `source_workbook`source_workbook=load_workbook(filename='fredgraph.xlsx')# an .xlsx workbook can have multiple sheets# print their names here for reference(source_workbook.sheetnames)# loop through the worksheets in `source_workbook`forsheet_num,sheet_nameinenumerate(source_workbook.sheetnames):# create a variable that points to the current worksheet by# passing the current value of `sheet_name` to `source_workbook`current_sheet=source_workbook[sheet_name]# print `sheet_name`, just to see what it is(sheet_name)# create an output file called "xlsx_"+sheet_nameoutput_file=open("xlsx_"+sheet_name+".csv","w")# use this csv library's "writer" recipe to easily write rows of data# to `output_file`, instead of reading data *from* itoutput_writer=csv.writer(output_file)# loop through every row in our sheetforrowincurrent_sheet.iter_rows():# we'll create an empty list where we'll put the actual# values of the cells in each rowrow_cells=[]# for every cell (or column) in each row....forcellinrow:# let's print what's in here, just to see how the code sees it(cell,cell.value)# add the values to the end of our list with the `append()` methodrow_cells.append(cell.value)# write our newly (re)constructed data row to the output fileoutput_writer.writerow(row_cells)# officially close the `.csv` file we just wrote all that data tooutput_file.close()
Like the csv library’s
DictReader()function,openpyxl’sload_workbook()function adds properties to our source data, in this case, one that shows us the names of all the data sheets in our workbook.
Even though our example workbook only includes one worksheet, we might have more in the future. We’ll use the
enumerate()function so we can access both an iterator and the sheet name. This will help us create one .csv file per worksheet.
Each sheet in our
source_workbookwill need its own, uniquely named output .csv file. To generate these, we’ll “open” a new file with the name"xlsx_"+sheet_name+".csv"and make it writable by passingwas the “mode” argument (up until now, we’ve used thermode to read data from .csvs).
The function
iter_rows()is specific to the openpyxl library. Here, it converts the rows ofsource_workbookinto a list that can be iterated, or looped, over.
The openpyxl library treats each data cell as a Python
tupledata type. If we try to just print the rows ofcurrent_sheetdirectly, we’ll get sort of unhelpful cell locations, rather than the data values they contain. To address this, we’ll make another loop inside this one to go through every cell in every row one at a time and add the actual data values torow_cells.
Notice that this code is left-aligned with the
for cell in rowcode in the example. This means that it is outside that loop and so will only be run after all the cells in a given row have been appended to our list.
This script also begins to demonstrate the way that, just as two chefs may have different ways of preparing the same dish, library creators may make different choices about how to (re)structure each source file type—with corresponding implications for our code. The creators of the openpyxl library, for example, chose to store each data cell’s location label (e.g., A6) and the value it contains in a Python tuple. That design decision is why we need a second for loop to go through each row of data—because we actually have to access the data cell by cell in order to build the Python list that will become a single row in our output .csv file. Likewise, if you use a spreadsheet program to open the xlsx_FRED Graph.csv created by the script in Example 4-4, you’ll see that the original .xls file shows the values in the observation_date column in a YYYY-MM-DD format, but our output file shows those values in a YYYY-MM-DD HH:MM:SS format. This is because the creator(s) of openpyxl decided that it would automatically convert any “date-like” data strings into the Python datetime datatype. Obviously, none of these choices are right or wrong; we simply need to account for them in writing our code so that we don’t distort or misinterpret the source data.
Now that we’ve wrangled the .xlsx version of our data file, let’s see what happens when we parse it as an .ods, as shown in Example 4-5.
Example 4-5. ods_parsing.py
# an example of reading data from an .ods file with Python, using the# "pyexcel_ods" library. First, you'll need to pip install the library:# https://pypi.org/project/pyexcel-ods/# specify the "chapter" of the "pyexcel_ods" library you want to import,# in this case, `get_data`frompyexcel_odsimportget_data# import the `csv` library, to create our output fileimportcsv# pass our filename as an ingredient to the `pyexcel_ods` library's# `get_data()` "recipe"# store the result in a variable called `source_workbook`source_workbook=get_data("fredgraph.ods")# an `.ods` workbook can have multiple sheetsforsheet_name,sheet_datainsource_workbook.items():# print `sheet_name`, just to see what it is(sheet_name)# create "ods_"+sheet_name+".csv" as an output file for the current sheetoutput_file=open("ods_"+sheet_name+".csv","w")# use this csv library's "writer" recipe to easily write rows of data# to `output_file`, instead of reading data *from* itoutput_writer=csv.writer(output_file)# now, we need to loop through every row in our sheetforrowinsheet_data:# use the `writerow` recipe to write each `row`# directly to our output fileoutput_writer.writerow(row)# officially close the `.csv` file we just wrote all that data tooutput_file.close()
The pyexcel_ods library converts our source data into Python’s
OrderedDictdata type. The associateditems()method then lets us access each sheet’s name and data as a key/value pair that we can loop through. In this case,sheet_nameis the “key” and the entire worksheet’s data is the “value.”
Here,
sheet_datais already a list, so we can just loop through that list with a basicforloop.
This library converts each row in a worksheet to a list, which is why we can pass these directly to the
writerow()method.
In the case of the pyexcel_ods library, the contents of our output .csv file much more closely resembles what we see visually when we open the original fredgraph.xls via a spreadsheet program like Google Sheets—the observation_date field, for example, is in a simple YYYY-MM-DD format. Moreover, the library creator(s) decided to treat the values in each row as a list, allowing us to write each record directly to our output file without creating any additional loops or lists.
Finally, let’s see what happens when we use the xlrd library to parse the original .xls file directly in Example 4-6.
Example 4-6. xls_parsing.py
# a simple example of reading data from a .xls file with Python# using the "xrld" library. First, pip install the xlrd library:# https://pypi.org/project/xlrd/2.0.1/# import the "xlrd" libraryimportxlrd# import the `csv` library, to create our output fileimportcsv# pass our filename as an ingredient to the `xlrd` library's# `open_workbook()` "recipe"# store the result in a variable called `source_workbook`source_workbook=xlrd.open_workbook("fredgraph.xls")# an `.xls` workbook can have multiple sheetsforsheet_nameinsource_workbook.sheet_names():# create a variable that points to the current worksheet by# passing the current value of `sheet_name` to the `sheet_by_name` recipecurrent_sheet=source_workbook.sheet_by_name(sheet_name)# print `sheet_name`, just to see what it is(sheet_name)# create "xls_"+sheet_name+".csv" as an output file for the current sheetoutput_file=open("xls_"+sheet_name+".csv","w")# use the `csv` library's "writer" recipe to easily write rows of data# to `output_file`, instead of reading data *from* itoutput_writer=csv.writer(output_file)# now, we need to loop through every row in our sheetforrow_num,rowinenumerate(current_sheet.get_rows()):# each row is already a list, but we need to use the `row_value()`# method to access them# then we can use the `writerow` recipe to write them# directly to our output fileoutput_writer.writerow(current_sheet.row_values(row_num))# officially close the `.csv` file we just wrote all that data tooutput_file.close()
Notice that this structure is similar to the one we use when working with the csv library.
The function
get_rows()is specific to the xlrd library; it converts the rows of our current worksheet into a list that can be looped over.12
There will be some funkiness within the “dates” written to our output file.13 We’ll look at how to fix up the dates in “Decrypting Excel Dates”.
One thing we’ll see in this output file is some serious weirdness in the values recorded in the observation_date field, reflecting the fact that, as the xlrd library’s creators put it:14
Dates in Excel spreadsheets: In reality, there are no such things. What you have are floating point numbers and pious hope.
As a result, getting a useful, human-readable date out of an .xls file requires some significant cleanup, which we’ll address in “Decrypting Excel Dates”.
As these exercises have hopefully demonstrated, with some clever libraries and a few tweaks to our basic code configuration, it’s possible to wrangle data from a wide range of table-type data formats with Python quickly and easily. At the same time, I hope that these examples have also illustrated why working with text-based and/or open source formats is almost always preferable,15 because they often require less “cleaning” and transformation to get them into a clear, usable state.
Finally, Fixed-Width
Though I didn’t mention it at the top of this section, one of the very oldest versions of table-type data is what’s known as “fixed-width.” As the name implies, each data column in a fixed-width table contains a specific, predefined number of characters—and always that number of characters. This means that the meaningful data in fixed-width files are often padded with extra characters, such as spaces or zeroes.
Though very uncommon in contemporary data systems, you are still likely to encounter fixed-width formats if you’re working with government data sources whose infrastructure may be decades old.16 For example, the US National Oceanic and Atmospheric Administration (NOAA), whose origins date back to the early 19th century, offers a wide range of detailed, up-to-date weather information online for free through its Global Historical Climatology Network, much of which is published in a fixed-width format. For example, information about the stations’ unique identifier, locations, and what network(s) they are a part of is stored in the ghcnd-stations.txt file. To interpret any actual weather data readings (many of which are also released as fixed-width files), you’ll need to cross-reference the station data with the weather data.
Even more than other table-type data files, working with fixed-width data can be especially tricky if you don’t have access to the metadata that describes how the file and its fields are organized. With delimited files, it’s often possible to eyeball the file in a text editor and identify the delimiter used with a reasonable level of confidence. At worst, you can simply try parsing the file using different delimiters and see which yields the best results. With fixed-width files—especially large ones—if there’s no data for a particular field in the sample of the data you inspect, it’s easy to end up inadvertently lumping together multiple data fields.
Fortunately, metadata about the ghcnd-stations.txt file that we’re using as our data source is included in the readme.txt file in the same folder on the NOAA site.
Looking through that readme.txt file, we find the heading IV. FORMAT OF "ghcnd-stations.txt", which contains the following table:
------------------------------ Variable Columns Type ------------------------------ ID 1-11 Character LATITUDE 13-20 Real LONGITUDE 22-30 Real ELEVATION 32-37 Real STATE 39-40 Character NAME 42-71 Character GSN FLAG 73-75 Character HCN/CRN FLAG 77-79 Character WMO ID 81-85 Character ------------------------------
This is followed by a detailed description of what each field contains or means, including information like units. Thanks to this robust data dictionary, we now know not just how the ghcnd-stations.txt file is organized but also how to interpret the information it contains. As we’ll see in Chapter 6, finding (or building) a data dictionary is an essential part of assessing or improving the quality of our data. At the moment, however, we can just focus on transforming this fixed-width file into a .csv, as detailed in Example 4-7.
Example 4-7. fixed_width_parsing.py
# an example of reading data from a fixed-width file with Python.# the source file for this example comes from NOAA and can be accessed here:# https://www1.ncdc.noaa.gov/pub/data/ghcn/daily/ghcnd-stations.txt# the metadata for the file can be found here:# https://www1.ncdc.noaa.gov/pub/data/ghcn/daily/readme.txt# import the `csv` library, to create our output fileimportcsvfilename="ghcnd-stations"# reading from a basic text file doesn't require any special libraries# so we'll just open the file in read format ("r") as usualsource_file=open(filename+".txt","r")# the built-in "readlines()" method does just what you'd think:# it reads in a text file and converts it to a list of linesstations_list=source_file.readlines()# create an output file for our transformed dataoutput_file=open(filename+".csv","w")# use the `csv` library's "writer" recipe to easily write rows of data# to `output_file`, instead of reading data *from* itoutput_writer=csv.writer(output_file)# create the header listheaders=["ID","LATITUDE","LONGITUDE","ELEVATION","STATE","NAME","GSN_FLAG","HCNCRN_FLAG","WMO_ID"]# write our headers to the output fileoutput_writer.writerow(headers)# loop through each line of our file (multiple "sheets" are not possible)forlineinstations_list:# create an empty list, to which we'll append each set of characters that# makes up a given "column" of datanew_row=[]# ID: positions 1-11new_row.append(line[0:11])# LATITUDE: positions 13-20new_row.append(line[12:20])# LONGITUDE: positions 22-30new_row.append(line[21:30])# ELEVATION: positions 32-37new_row.append(line[31:37])# STATE: positions 39-40new_row.append(line[38:40])# NAME: positions 42-71new_row.append(line[41:71])# GSN_FLAG: positions 73-75new_row.append(line[72:75])# HCNCRN_FLAG: positions 77-79new_row.append(line[76:79])# WMO_ID: positions 81-85new_row.append(line[80:85])# now all that's left is to use the# `writerow` function to write new_row to our output fileoutput_writer.writerow(new_row)# officially close the `.csv` file we just wrote all that data tooutput_file.close()
Since we don’t have anything within the file that we can draw on for column headers, we have to “hard code” them based on the information in the readme.txt file. Note that I’ve eliminated special characters and used underscores in place of spaces to minimize hassles when cleaning and analyzing this data later on.
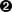
Python actually views lines of text as just lists of characters, so we can just tell it to give us the characters between two numbered index positions. Like the
range()function, the character at the first position is included, but the second number is not. Also recall that Python starts counting lists of items at zero (often called zero-indexing). This means that for each entry, the first number will be one less than whatever the metadata says, but the righthand number will be the same.
If you run the script in Example 4-7 and open your output .csv file in a spreadsheet program, you’ll notice that the values in some of the columns are not formatted consistently. For example, in the ELEVATION column, the numbers with decimals are left justified, but those without decimals are right justified. What’s going on?
Once again, opening the file in a text editor is enlightening. Although the file we’ve created is technically comma separated, the values we put into each of our newly “delimited” columns still contain the extra spaces that existed in the original file. As a result, our new file still looks pretty “fixed-width.”
In other words—just as we saw in the case of Excel “dates”—converting our file to a .csv does not “automagically” generate sensible data types in our output file. Determining what data type each field should have—and cleaning them up so that they behave appropriately—is part of the data cleaning process that we’ll address in Chapter 7.
Feed-Based Data—Web-Driven Live Updates
The structure of table-type data formats is well suited to a world where most “data” has already been filtered, revised, and processed into a relatively well-organized collection of numbers, dates, and short strings. With the rise of the internet, however, came the need to transmit large quantities of the type of “free” text found in, for example, news stories and social media feeds. Because this type of data content typically includes characters like commas, periods, and quotation marks that affect its semantic meaning, fitting it into a traditional delimited format will be problematic at best. What’s more, the horizontal bias of delimited formats (which involves lots of left-right scrolling) runs counter to the vertical-scrolling conventions of the web. Feed-based data formats have been designed to address both of these limitations.
At a high level, there are two main types of feed-based data formats: XML and JSON. Both are text-based formats that allow the data provider to define their own unique data structure, making them extremely flexible and, consequently, useful for the wide variety of content found on internet-connected websites and platforms. Whether they’re located online or you save a copy locally, you’ll recognize these formats, in part, by their coordinating .xml and .json file extensions:
- .xml
-
Extensible Markup Language encompasses a broad range of file formats, including .rss, .atom, and even .html. As the most generic type of markup language, XML is extremely flexible and was perhaps the original data format for web-based data feeds.
- .json
-
JavaScript Object Notation files are somewhat more recent than XML files but serve a similar purpose. In general, JSON files are less descriptive (and therefore shorter and more concise) than XML files. This means that they can encode an almost identical amount of data as an XML file while taking up less space, which is especially important for speed on the mobile web. Equally important is the fact that JSON files are essentially large
objectdata types within the JavaScript programming language—which is the language that underpins many, if not most, websites and mobile apps. This means that parsing JSON-formatted data is very easy for any site or program that uses JavaScript, especially when compared with XML. Fortunately, JavaScriptobjectdata types are very similar to Pythondictdata types, which also makes working with JSON in Python very straightforward.
Before we dive into how to work with each of these file types in Python, let’s review when we might want feed-type data and where to find it when we do.
When to work with feed-type data
In a sense, feed-type data is to the 21st century what table-type data was to the 20th: the sheer volume of feed-type data generated, stored, and exchanged on the web every day is probably millions of times greater than that of all of the table-type data in the world put together—in large part because feed-type data is what powers social media sites, news apps, and everything in between.
From a data wrangling perspective, you’ll generally want feed-type data when the phenomenon you’re exploring is time sensitive and updated on a frequent and/or unpredictable basis. Typically, this type of data is generated in response to a human or natural process, such as (once again) posting to social media, publishing a news story, or recording an earthquake.
Both file-based, table-type data and web-based, feed-type data can contain historical information, but as we discussed at the start of this chapter, the former usually reflects the data as it stood at a fixed point in time. The latter, by contrast, is typically organized in a “reverse-chronological” (most recent first) order, with the first entry being whatever data record was most recently created at the time you accessed the data, rather than a predetermined publication date.
Where to find feed-type data
Feed-type data is found almost exclusively on the web, often at special URLs known as application programming interface (API) endpoints. We’ll get into the details of working with APIs in Chapter 5, but for now all you need to know is that API endpoints are really just data-only web pages: you can view many of them using a regular web browser, but all you’ll see is the data itself. Some API endpoints will even return different data depending on the information you send to them, and this is part of what makes working with feed-type data so flexible: by changing just a few words or values in your code, you can access a totally different dataset!
Finding APIs that offer feed-type data doesn’t require too much in the way of special search strategies because usually the sites and services that have APIs want you to find them. Why? Simply put, when someone writes code that makes use of an API, it (usually) returns some benefit to the company that provides it—even if that benefit is just more public exposure. In the early days of Twitter, for example, many web developers wrote programs using the Twitter API—both making the platform more useful and saving the company the expense and effort of figuring out what users wanted and then building it. By making so much of their platform data available for free (at first), the API gave rise to several companies that Twitter would eventually purchase—though many more would also be put out of business when either the API or its terms of service changed.17 This highlights one of the particular issues that can arise when working with any type of data, but especially the feed-type data made available by for-profit companies: both the data and your right to access it can change at any time, without warning. So while feed-type data sources are indeed valuable, they are also ephemeral in more ways than one.
Wrangling Feed-Type Data with Python
As with table-type data, wrangling feed-type data in Python is made possible by a combination of helpful libraries and the fact that formats like JSON already resemble existing data types in the Python programming language. Moreover, we’ll see in the following sections that XML and JSON are often functionally interchangeable for our purposes (though many APIs will only offer data in one format or the other).
XML: One markup to rule them all
Markup languages are among the oldest forms of standardized document formats in computing, designed with the goal of creating text-based documents that can be easily read by both humans and machines. XML became an increasingly important part of internet infrastructure in the 1990s as the variety of devices accessing and displaying web-based information made the separation of content (e.g., text and images) from formatting (e.g., page layout) more of a necessity. Unlike an HTML document—in which content and formatting are fully commingled—an XML document says pretty much nothing about how its information should be displayed. Instead, its tags and attributes act as metadata about what kind of information it contains, along with the data itself.
To get a feel for what XML looks like, take a look at Example 4-8.
Example 4-8. A sample XML document
<?xml version="1.0" encoding="UTF-8"?><mainDoc><!--This is a comment--><elements><element1>This is some text in the document.</element1><element2>This is some other data in the document.</element2><element3someAttribute="aValue"/></elements><someElementanAttribute="anotherValue">More content</someElement></mainDoc>
There are a couple of things going here. The very first line is called the document type (or doc-type) declaration; it’s letting us know that the rest of the document should be interpreted as XML (as opposed to any of the other web or markup languages, some of which we’ll review later in this chapter).
Starting with the line:
<mainDoc>
we are into the substance of the document itself. Part of what makes XML so flexible is that it only contains two real grammatical structures, both of which are included in Example 4-8:
- tags
-
Tags can be either paired (like
element1,element2,someElement, or evenmainDoc) or self-closed (likeelement3). The name of a tag is always enclosed by carets (<>). In the case of a closing tag, the opening caret is immediately followed by a forward slash (/). A matched pair of tags, or a self-closed tag, are also described as XML elements. - attributes
-
Attributes can exist only inside of tags (like
anAttribute). Attributes are a type of key/value pair in which the attribute name (or key) is immediately followed by an equals sign (=), followed by the value surrounded by double quotation marks ("").
An XML element is whatever is contained between an opening tag and its matching closing tag (e.g., <elements> and </elements>). As such, a given XML element may contain many tags, each of which may also contain other tags. Any tags may also have any number of attributes (including none). A self-closed tag is also considered an element.
The only other meaningful rule for structuring XML documents is that when tags appear inside other tags, the most recently opened tag must be closed first. In other words, while this is a legitimate XML structure:
<outerElement><!-- Notice that that the `innerElement1` is closedbefore the `innerElement2` tag is opened --><innerElement1>Some content</innerElement1><innerElement2>More content</innerElement2></outerElement>
this is not:
<outerElement><!-- NOPE! The `innerElement2` tag was openedbefore the `innerElement1` tag was closed --><innerElement1>Some content<innerElement2>More content</innerElement1></innerElement2></outerElement>
This principle of last opened, first closed is also described as nesting, similar to the “nested” for...in loops from Figure 2-3.18 Nesting is especially important in XML documents because it governs one of the primary mechanisms that we use to read or parse XML (and other markup language) documents with code. In an XML document, the first element after the doc-type declaration is known as the root element. If the XML document has been formatted, the root element will always be left justified, and any element that is nested directly within that element will be indented one level to the right and is referred to as a child element. In Example 4-8, then, <mainDoc> would be considered the root element, and <elements> would be its child. Likewise, <mainDoc> is the parent element of <elements> (Example 4-9).
Example 4-9. An annotated XML document
<?xml version="1.0" encoding="UTF-8"?><mainDoc><!--`mainDoc` is the *root* element, and `elements` is its *child*--><elements><!-- `elements` is the *parent* of `element1`, `element2`, and`element3`, which are *siblings* of one another --><element1>This is text data in the document.</element1><element2>This is some other data in the document.</element2><element3someAttribute="aValue"/></elements><!-- `someElement` is also a *child* of `mainDoc`,and a *sibling* of `elements` --><someElementanAttribute="anotherValue">More content</someElement></mainDoc>
Given this trend for genealogical jargon, you might be wondering: if <elements> is the parent of <element3>, and <mainDoc> is the parent of <elements>, does that make <mainDoc> the grandparent of <element3>? The answer is: yes, but no. While <mainDoc> is the “parent” of the “parent” of <element3>, the term “grandparent” is never used in describing an XML structure—that could get complicated fast! Instead, we simply describe the relationship as exactly that: <mainDoc> is the parent of the parent of <element3>.
Fortunately, there is no such complexity associated with XML attributes: they are simply key/value pairs, and they can only exist within XML tags, like so:
<element3someAttribute="aValue"/>
Note that there is no space on either side of the equals sign, just as there is no space between the carets and slashes of an element tag.
Like writing in English (or in Python), the question of when to use tags versus attributes for a particular piece of information is largely a matter of preference and style. Both Examples 4-10 and 4-11, for example, contain the same information about this book, but each is structured slightly differently.
Example 4-10. Sample XML book data—more attributes
<aBook><bookURLurl="https://www.oreilly.com/library/view/practical-python-data/9781492091493"/><bookAbstract>There are awesome discoveries to be made and valuable stories to be told in datasets--and this book will help you uncover them.</bookAbstract><pubDatedate="2022-02-01"/></aBook>
Example 4-11. Sample XML book data—more elements
<aBook><bookURL>https://www.oreilly.com/library/view/practical-python-data/9781492091493</bookURL><bookAbstract>There are awesome discoveries to be made and valuable stories to be told in datasets--and this book will help you uncover them.</bookAbstract><pubDate>2022-02-01</pubDate></aBook>
This degree of flexibility means XML is highly adaptable to a wide variety of data sources and formatting preferences. At the same time, it can easily create a situation where every new data source requires writing custom code. Obviously, this would be a pretty inefficient system, especially if many people and organizations were publishing pretty similar types of data.
It’s not surprising, then, that there are a large number of XML specifications that define additional rules for formatting XML documents that are intended to hold particular types of data. I’m highlighting a few notable examples here, as these are formats you may come across in the course of your data wrangling work. Despite their various format names and file extensions, however, we can parse them all using the same method that we’ll look at shortly in Example 4-12:
- RSS
-
Really Simple Syndication is an XML specification first introduced in the late 1990s for news information. The .atom XML format is also widely used for these purposes.
- KML
-
Keyhole Markup Language is an internationally accepted standard for encoding both two-dimensional and three-dimensional geographic data and is compatible with tools like Google Earth.
- SVG
-
Scalable Vector Graphics is a commonly used format for graphics on the web, thanks to its ability to scale drawings without loss of quality. Many common graphics programs can output .svg files, which can then be included in web pages and other documents that will look good on a wide variety of screen sizes and devices.
- EPUB
-
Electronic publication format (.epub) is the widely accepted open standard for digital book publishing.
As you can see from the preceding list, some common XML formats clearly indicate their relationship to XML; many others do not.19
Now that we have a high-level sense of how XML files work, let’s see what it takes to parse one with Python. Although Python has some built-in tools for parsing XML, we’ll be using a library called lxml, which is particularly good at quickly parsing large XML files. Even though our example files that follow are quite small, know that we could use basically the same code even if our data files got considerably larger.
To begin with, we’ll be using an XML version of the same “U6” unemployment data that I’ve already downloaded from the FRED website using its API.20 After downloading a copy of this file from Google Drive, you can use the script in Example 4-12 to convert the source XML to a .csv.
Start with the pip install:
pip install lxml
Example 4-12. xml_parsing.py
# an example of reading data from an .xml file with Python, using the "lxml"# library.# first, you'll need to pip install the lxml library:# https://pypi.org/project/lxml/# a helpful tutorial can be found here: https://lxml.de/tutorial.html# the data used here is an instance of# https://api.stlouisfed.org/fred/series/observations?series_id=U6RATE& \# api_key=YOUR_API_KEY_HERE# specify the "chapter" of the `lxml` library you want to import,# in this case, `etree`, which stands for "ElementTree"fromlxmlimportetree# import the `csv` library, to create our output fileimportcsv# choose a filenamefilename="U6_FRED_data"# open our data file in read format, using "rb" as the "mode"xml_source_file=open(filename+".xml","rb")# pass our xml_source_file as an ingredient to the `lxml` library's# `etree.parse()` method and store the result in a variable called `xml_doc`xml_doc=etree.parse(xml_source_file)# start by getting the current xml document's "root" elementdocument_root=xml_doc.getroot()# let's print it out to see what it looks like(etree.tostring(document_root))# confirm that `document_root` is a well-formed XML elementifetree.iselement(document_root):# create our output file, naming it "xml_"+filename+".csvoutput_file=open("xml_"+filename+".csv","w")# use the `csv` library's "writer" recipe to easily write rows of data# to `output_file`, instead of reading data *from* itoutput_writer=csv.writer(output_file)# grab the first element of our xml document (using `document_root[0]`)# and write its attribute keys as column headers to our output fileoutput_writer.writerow(document_root[0].attrib.keys())# now, we need to loop through every element in our XML fileforchildindocument_root:# now we'll use the `.values()` method to get each element's values# as a list and then use that directly with the `writerow` recipeoutput_writer.writerow(child.attrib.values())# officially close the `.csv` file we just wrote all that data tooutput_file.close()
In this instance, there’s nothing within the data file (like a sheet name) that we can pull as a filename, so we’ll just make our own and use it to both load our source data and label our output file.
I lied! The values we’ve been using for the “mode” of the
open()function assume we want to interpret the source file as text. But because the lxml library expects byte data rather than text, we’ll userb(“read bytes”) as the “mode.”
There is a lot of malformed XML out there! In order to make sure that what looks like good XML actually is, we’ll retrieve the current XML document’s “root” element and make sure that it works.

Because our XML is currently stored as byte data, we need to use the
etree.tostring()method in order to view it as one.
Thanks to the lxml, each XML element (or “node”) in our document has a property called
attrib, whose data type is a Python dictionary (dict). Using the.keys()method returns all of our XML element’s attribute keys as a list. Since all of the elements in our source file are identical, we can use the keys of the first one to create a “header” row for our output file.
The lxml library converts XML elements to lists, so we can use a simple
for...inloop to go through the elements in our document.
As it happens, the XML version of our unemployment data is structured very simply: it’s just a list of elements, and all the values we want to access are stored as attributes. As a result, we were able to pull the attribute values out of each element as a list and write them directly to our .csv file with only one line of code.
Of course, there are many times when we’ll want to pull data from more complex XML formats, especially those like RSS or Atom. To see what it takes to handle something slightly more complex, in Example 4-13 we’ll parse the BBC’s RSS feed of science and environment stories, which you can download a copy of from my Google Drive.
Example 4-13. rss_parsing.py
# an example of reading data from an .xml file with Python, using the "lxml"# library.# first, you'll need to pip install the lxml library:# https://pypi.org/project/lxml/# the data used here is an instance of# http://feeds.bbci.co.uk/news/science_and_environment/rss.xml# specify the "chapter" of the `lxml` library you want to import,# in this case, `etree`, which stands for "ElementTree"fromlxmlimportetree# import the `csv` library, to create our output fileimportcsv# choose a filename, for simplicityfilename="BBC News - Science & Environment XML Feed"# open our data file in read format, using "rb" as the "mode"xml_source_file=open(filename+".xml","rb")# pass our xml_source_file as an ingredient to the `lxml` library's# `etree.parse()` method and store the result in a variable called `xml_doc`xml_doc=etree.parse(xml_source_file)# start by getting the current xml document's "root" elementdocument_root=xml_doc.getroot()# if the document_root is a well-formed XML elementifetree.iselement(document_root):# create our output file, naming it "rss_"+filename+".csv"output_file=open("rss_"+filename+".csv","w")# use the `csv` library's "writer" recipe to easily write rows of data# to `output_file`, instead of reading data *from* itoutput_writer=csv.writer(output_file)# document_root[0] is the "channel" elementmain_channel=document_root[0]# the `find()` method returns *only* the first instance of the element namearticle_example=main_channel.find('item')# create an empty list in which to store our future column headerstag_list=[]forchildinarticle_example.iterdescendants():# add each tag to our would-be header listtag_list.append(child.tag)# if the current tag has any attributesifchild.attrib:# loop through the attribute keys in the tagforattribute_nameinchild.attrib.keys():# append the attribute name to our `tag_list` column headerstag_list.append(attribute_name)# write the contents of `tag_list` to our output file as column headersoutput_writer.writerow(tag_list)# now we want to grab *every* <item> element in our file# so we use the `findall()` method instead of `find()`foriteminmain_channel.findall('item'):# empty list for holding our new row's contentnew_row=[]# now we'll use our list of tags to get the contents of each elementfortagintag_list:# if there is anything in the element with a given tag nameifitem.findtext(tag):# append it to our new rownew_row.append(item.findtext(tag))# otherwise, make sure it's the "isPermaLink" attributeeliftag=="isPermaLink":# grab its value from the <guid> element# and append it to our rownew_row.append(item.find('guid').get("isPermaLink"))# write the new row to our output file!output_writer.writerow(new_row)# officially close the `.csv` file we just wrote all that data tooutput_file.close()
As always, we’ll want to balance what we handle programmatically and what we review visually. In looking at our data, it’s clear that each article’s information is stored in a separate
itemelement. Since copying over the individual tag and attribute names would be time-consuming and error prone, however, we’ll go through oneitemelement and make a list of all the tags (and attributes) within it, which we’ll then use as the column headers for our output .csv file.
The
iterdescendants()method is particular to the lxml library. It returns only the descendants of an XML element, while the more commoniter()method would return both the element itself and its children or “descendants.”
Using
child.tagwill retrieve the text of the tagname of the child element. For example, for the<pubDate>`element it will returnpubDate.
Only one tag in our
<item>element has an attribute, but we still want to include it in our output.
The
keys()method will give us a list of all the keys in the list of attributes that belong to the tag. Be sure to get its name as a string (instead of a one-item list).
That whole
article_exampleforloop was just to buildtag_list—but it was worth it!
As you can see from Example 4-13, with the help of the lxml library, parsing even slightly more complex XML in Python is still reasonably straightforward.
While XML is still a popular data format for news feeds and a handful of other file types, there are a number of features that make it less than ideal for handling the high-volume data feeds of the modern web.
First, there is the simple issue of size. While XML files can be wonderfully descriptive—reducing the need for separate data dictionaries—the fact that most elements contain both an opening tag and a corresponding closing tag (e.g., <item> and </item>) also makes XML somewhat verbose: there is a lot of text in an XML document that isn’t content. This isn’t a big deal when your document has a few dozen or even a few thousand elements, but when you’re trying to handle millions or billions of posts on the social web, all that redundant text can really slow things down.
Second, while XML isn’t exactly difficult to transform into other data formats, the process also isn’t exactly seamless. The lxml library (among others) makes parsing XML with Python pretty simple, but doing the same task with web-focused languages like JavaScript is convoluted and onerous. Given JavaScript’s prevalence on the web, it’s not surprising that a feed-type data format that works seamlessly with JavaScript would be developed at some point. As we’ll see in
the next section,
many of XML’s limitations as a data format are addressed by the object-like nature of the .json format, which is at this point the most popular format for feed-type data on the internet.
JSON: Web data, the next generation
In principle, JSON is similar to XML in that it uses nesting to cluster related pieces of information into records and fields. JSON is also fairly human readable, though the fact that it doesn’t support comments means that JSON feeds may require more robust data dictionaries than XML documents.
To get started, let’s take a look at the small JSON document in Example 4-14.
Example 4-14. Sample JSON document
{"author":"Susan E. McGregor","book":{"bookURL":"https://www.oreilly.com/library/view/practical-python-data/9781492091493/","bookAbstract":"There are awesome discoveries to be made and valuablestories to be told in datasets--and this book will help you uncoverthem.","pubDate":"2022-02-01"},"papers":[{"paperURL":"https://www.usenix.org/conference/usenixsecurity15/technical-sessions/presentation/mcgregor","paperTitle":"Investigating the computer security practices and needsof journalists","pubDate":"2015-08-12"},{"paperURL":"https://www.aclweb.org/anthology/W18-5104.pdf","paperTitle":"Predictive embeddings for hate speech detection ontwitter","pubDate":"2018-10-31"}]}
Like XML, the grammatical “rules” of JSON are quite simple: there are only three distinct data structures in JSON documents, all of which appear in Example 4-14:
- Key/value pairs
-
Technically, everything within a JSON document is a key/value pair, with the key enclosed in quotes to the left of a colon (
:) and the value being whatever appears to the right of the colon. Note that while keys must always be strings, values can be strings (as inauthor), objects (as inbook), or lists (as inpapers). - Objects
-
These are opened and closed using pairs of curly braces (
{}). In Example 4-14, there are four objects total: the document itself (indicated by the left-justified curly braces), thebookobject, and the two unnamed objects in thepaperslist. - Lists
-
These are enclosed by square brackets (
[]) and can only contain comma-separated objects.
While XML and JSON can be used to encode the same data, there are some notable differences in what each allows. For example, JSON files do not contain a doc-type specification, nor can they include comments. Also, while XML lists are somewhat implicit (any repeated element functions something like a list), in JSON, lists must be specified by square brackets ([]).
Finally, although JSON was designed with JavaScript in mind, you may have noticed that its structures are very similar to Python dict and list types. This is part of what makes parsing JSON very straightforward with Python as well as JavaScript (and a range of other languages).
To see just how straightforward this is, in Example 4-15 we’ll parse the same data as we did in Example 4-12 but in the .json format also provided by the FRED API. You can download the file from this Google Drive link: https://drive.google.com/file/d/1Mpb2f5qYgHnKcU1sTxTmhOPHfzIdeBsq/view?usp=sharing.
Example 4-15. json_parsing.py
# a simple example of reading data from a .json file with Python,# using the built-in "json" library. The data used here is an instance of# https://api.stlouisfed.org/fred/series/observations?series_id=U6RATE& \# file_type=json&api_key=YOUR_API_KEY_HERE# import the `json` library, since that's our source file formatimportjson# import the `csv` library, to create our output fileimportcsv# choose a filenamefilename="U6_FRED_data"# open the file in read format ("r") as usualjson_source_file=open(filename+".json","r")# pass the `json_source_file` as an ingredient to the json library's `load()`# method and store the result in a variable called `json_data`json_data=json.load(json_source_file)# create our output file, naming it "json_"+filenameoutput_file=open("json_"+filename+".csv","w")# use the `csv` library's "writer" recipe to easily write rows of data# to `output_file`, instead of reading data *from* itoutput_writer=csv.writer(output_file)# grab the first element (at position "0"), and use its keys as the column headersoutput_writer.writerow(list(json_data["observations"][0].keys()))forobjinjson_data["observations"]:# we'll create an empty list where we'll put the actual values of each objectobj_values=[]# for every `key` (which will become a column), in each objectforkey,valueinobj.items():# let's print what's in here, just to see how the code sees it(key,value)# add the values to our listobj_values.append(value)# now we've got the whole row, write the data to our output fileoutput_writer.writerow(obj_values)# officially close the `.csv` file we just wrote all that data tooutput_file.close()
Because the json library interprets every object as a dictionary view object, we need to tell Python to convert it to a regular list using the
list()function.
In most cases, the simplest way to find the name (or “key”) of the main JSON object in our document is just to look at it. Because JSON data is often rendered on a single line, however, we can get a better sense of its structure by pasting it into JSONLint. This lets us see that our target data is a list whose key is
observations.
Because of the way that the json library works, if we try to just write the rows directly, we’ll get the values labeled with
dict, rather than the data values themselves. So we need to make another loop that goes through every value in everyjsonobject one at a time and appends that value to ourobj_valueslist.
Although JSON is not quite as human readable as XML, it has other advantages that we’ve touched on, like smaller file size and broader code compatibility. Likewise, while JSON is not as descriptive as XML, JSON data sources (often APIs) are usually reasonably well documented; this reduces the need to simply infer what a given key/value pair is describing. Like all work with data, however, the first step in wrangling JSON-formatted data is to understand its context as much as possible.
Working with Unstructured Data
As we discussed in “Structured Versus Unstructured Data”, the process of creating data depends on introducing some structure to information; otherwise we can’t methodically analyze or derive meaning from it. Even though the latter often includes large segments of human-written “free” text, both table-type and feed-type data are relatively structured and, most importantly, machine readable.
When we deal with unstructured data, by contrast, our work always involves approximations: we cannot be certain that our programmatic wrangling efforts will return an accurate interpretation of the underlying information. This is because most unstructured data is a representation of content that is designed to be perceived and interpreted by humans. And, as we discussed in Chapter 2, while they can process large quantities of data much faster and with fewer errors than humans can, computers can still be tricked by unstructured data that would never fool a human, such as mistaking a slightly modified stop sign for a speed limit sign. Naturally, this means that when dealing with data that isn’t machine readable, we always need to do extra proofing and verification—but Python can still help us wrangle such data into a more usable format.
Image-Based Text: Accessing Data in PDFs
The Portable Document Format (PDF) was created in the early 1990s as a mechanism for preserving the visual integrity of electronic documents—whether they were created in a text-editing program or captured from printed materials.22 Preserving documents’ visual appearance also meant that, unlike machine-readable formats (such as word-processing documents), it was difficult to alter or extract their contents—an important feature for creating everything from digital versions of contracts to formal letters.
In other words, wrangling the data in PDFs was, at first, somewhat difficult by design. Because accessing the data in printed documents is a shared problem, however, work in optical character recognition (OCR) actually began as early as the late 19th century.23 Even digital OCR tools have been widely available in software packages and online for decades, so while they are far from perfect, the data contained in this type of file is also not entirely out of reach.
When to work with text in PDFs
In general, working with PDFs is a last resort (much, as we’ll see in Chapter 5, as web scraping should be). In general, if you can avoid relying on PDF information, you should. As noted previously, the process of extracting information from PDFs will generally yield an approximation of the document’s contents, so proofing for accuracy is a nonnegotiable part of any .pdf-based data wrangling workflow. That said, there is an enormous quantity of information that is only available as images or PDFs of scanned documents, and Python is an efficient way to extract a reasonably accurate first version of such document text.
Where to find PDFs
If you’re confident that the data you want can only be found in PDF format, then you can (and should) use the tips in “Smart Searching for Specific Data Types” to locate this file type using an online search. More than likely, you will request information from a person or organization, and they will provide it as PDFs, leaving you to deal with the problem of how to extract the information you need. As a result, most of the time you will not need to go looking for PDFs—more often than not they will, unfortunately, find you.
Wrangling PDFs with Python
Because PDFs can be generated both from machine-readable text (such as word-processing documents) and from scanned images, it is sometimes possible to extract the document’s “live” text programmatically with relatively few errors. While it seems straightforward, however, this method can still be unreliable because .pdf files can be generated with a wide range of encodings that can be difficult to detect accurately. So while this can be a high-accuracy method of extracting text from a .pdf, the likelihood of it working for any given file is low.
Because of this, I’m going to focus here on using OCR to recognize and extract the text in .pdf files. This will require two steps:
-
Convert the document pages into individual images.
-
Run OCR on the page images, extract the text, and write it to individual text files.
Unsurprisingly, we’ll need to install quite a few more Python libraries in order to make this all possible. First, we’ll install a couple of libraries for converting our .pdf pages to images. The first is a general-purpose library called poppler that is needed to make our Python-specific library pdf2image work. We’ll be using pdf2image to (you guessed it!) convert our .pdf file to a series of images:
sudo apt install poppler-utils
Then:
pip install pdf2image
Next, we need to install the tools for performing the OCR process. The first one is a general library called tesseract-ocr, which uses machine learning to recognize the text in images; the second is a Python library that relies on tesseract-ocr called pytesseract:
sudo apt-get install tesseract-ocr
Then:
pip install pytesseract
Finally, we need a helper library for Python that can do the computer vision needed to bridge the gap between our page images and our OCR library:
pip install opencv-python
Phew! If that seems like a lot of extra libraries, keep in mind that what we’re technically using here are is machine learning, one of those buzzy data science technologies that drives so much of the “artificial intelligence” out there. Fortunately for us, Tesseract in particular is relatively robust and inclusive: though it was originally developed by Hewlett-Packard as a proprietary system in the early 1980s, it was open sourced in 2005 and currently supports more than 100 languages—so feel free to try the solution in Example 4-16 out on non-English text as well!
Example 4-16. pdf_parsing.py
# a basic example of reading data from a .pdf file with Python,# using `pdf2image` to convert it to images, and then using the# openCV and `tesseract` libraries to extract the text# the source data was downloaded from:# https://files.stlouisfed.org/files/htdocs/publications/page1-econ/2020/12/01/ \# unemployment-insurance-a-tried-and-true-safety-net_SE.pdf# the built-in `operating system` or `os` Python library will let us create# a new folder in which to store our converted images and output textimportos# we'll import the `convert_from_path` "chapter" of the `pdf2image` libraryfrompdf2imageimportconvert_from_path# the built-in `glob`library offers a handy way to loop through all the files# in a folder that have a certain file extension, for exampleimportglob# `cv2` is the actual library name for `openCV`importcv2# and of course, we need our Python library for interfacing# with the tesseract OCR processimportpytesseract# we'll use the pdf name to name both our generated images and text filespdf_name="SafetyNet"# our source pdf is in the same folder as our Python scriptpdf_source_file=pdf_name+""# as long as a folder with the same name as the pdf does not already existifos.path.isdir(pdf_name)==False:# create a new folder with that nametarget_folder=os.mkdir(pdf_name)# store all the pages of the PDF in a variablepages=convert_from_path(pdf_source_file,300)# loop through all the converted pages, enumerating them so that the page# number can be used to label the resulting imagesforpage_num,pageinenumerate(pages):# create unique filenames for each page image, combining the# folder name and the page numberfilename=os.path.join(pdf_name,"p"+str(page_num)+".png")# save the image of the page in systempage.save(filename,'PNG')# next, go through all the files in the folder that end in `.png`forimg_fileinglob.glob(os.path.join(pdf_name,'*.png')):# replace the slash in the image's filename with a dottemp_name=img_file.replace("/",".")# pull the unique page name (e.g. `p2`) from the `temp_name`text_filename=temp_name.split(".")[1]# now! create a new, writable file, also in our target folder, that# has the same name as the image, but is a `.txt` fileoutput_file=open(os.path.join(pdf_name,text_filename+".txt"),"w")# use the `cv2` library to interpret our imageimg=cv2.imread(img_file)# create a new variable to hold the results of using pytesseract's# `image_to_string()` function, which will do just thatconverted_text=pytesseract.image_to_string(img)# write our extracted text to our output fileoutput_file.write(converted_text)# close the output fileoutput_file.close()
Here we pass the path to our source file (in this case, that is just the filename, because it is in the same folder as our script) and the desired dots per inch (DPI) resolution of the output images to the
convert_from_path()function. While setting a lower DPI will be much faster, the poorer-quality images may yield significantly less accurate OCR results. 300 DPI is a standard “print” quality resolution.
Here we use the os library’s
.joinfunction to save the new files into our target folder. We also have to use thestr()function to convert the page number into a string for use in the filename.
Note that
*.pngcan be translated to “any file ending in .png.” Theglob()function creates a list of all the filenames in the folder where our images are stored (which in this case has the valuepdf_name).
String manipulation is fiddly! To generate unique (but matching!) filenames for our OCR’d text files, we need to pull a unique page name out of
img_filewhose value starts withSafetyNet/and ends in.png. So we’ll replace the forward slash with a period to get something likeSafetyNet.p1.png, and then if wesplit()that on the period, we’ll get a list like:["SafetyNet", "p1", "png"]. Finally, we can access the “page name” at position 1. We need to do all this because we can’t be sure thatglob()will, for example, pullp1.pngfrom the image folder first, or that it will pull the images in order at all.
For the most part, running this script serves our purposes: with a few dozen lines of code, it converts a multipage PDF file first into images and then writes (most of) the contents to a series of new text files.
This all-in-one approach also has its limitations, however. Converting a PDF to images—or images to text—is the kind of task that we might need to do quite often but not always at the same time. In other words, it would probably be much more useful in the long run to have two separate scripts for solving this problem, and then to run them one after the other. In fact, with a little bit of tweaking, we could probably break up the preceding script in such a way that we could convert any PDF to images or any images to text without having to write any new code at all. Sounds pretty nifty, right?
This process of rethinking and reorganizing working code is known as code refactoring. In writing English, we would describe this as revising or editing, and the objective in both cases is the same: to make your work simpler, clearer, and more effective. And just like documentation, refactoring is actually another important way to scale your data wrangling work, because it makes reusing your code much more straightforward. We’ll look at various strategies for code refactoring and script reuse in Chapter 8.
Accessing PDF Tables with Tabula
If you looked at the text files produced in the preceding section, you’ll likely have noticed that there are a lot of “extras” in those files: page numbers and headers, line breaks, and other “cruft.” There are also some key elements missing, like images and tables.
While our data work won’t extend to analyzing images (that is a much more specialized area), it’s not unusual to find tables inside PDFs that hold data we might want to work with. In fact, this problem is common enough in my home field of journalism that a group of investigative journalists designed and built a tool called Tabula specifically to deal with this problem.
Tabula isn’t a Python library—it’s actually a standalone piece of software. To try it out, download the installer for your system; if you’re on a Chromebook or Linux machine, you’ll need to download the .zip file and follow the directions in README.txt. Whatever system you’re using, you’ll probably need to install the Java programming library first, which you can do by running the following command in a terminal window:
sudo apt install default-jre
Like some of the other open source tools we’ll discuss in later chapters (like OpenRefine, which I used to prepare some of the sample data in Chapter 2 and cover briefly in Chapter 11), Tabula does its work behind the scenes (though some of it is visible in the terminal window), and you interact with it in a web browser. This is a way to get access to a more traditional graphical interface while still leaving most of your computer’s resources free to do the heavy-duty data work.
Conclusion
Hopefully, the coding examples in this chapter have started to give you an idea of the wide variety of data wrangling problems you can solve with relatively little Python code, thanks to a combination of carefully selected libraries and those few essential Python scripting concepts that were introduced in Chapter 2.
You may also have noticed that, with the exception of our PDF text, the output of all the exercises in this chapter was essentially a .csv file. This is not by accident. Not only are .csv files efficient and versatile, but it turns out that to do almost any basic statistical analyses or visualizations, we need table-type data to work with. That’s not to say that it isn’t possible to analyze nontable data; that, in fact, is what much of contemporary computer science research (like machine learning) is all about. However, because those systems are often both complex and opaque, they’re not really suitable for the type of data wrangling work we’re focused on here. As such, we’ll spend our energy on the types of analyses that can help us understand, explain, and communicate new insights about the world.
Finally, while our work in this chapter focused on file-based data and pre-saved versions of feed-based data, in Chapter 5 we’ll explore how we can use Python with APIs and web scraping to wrangle data out of online systems and, where necessary, right off of web pages themselves!
1 Contrast these with some others you might know, like mp4 or png, which are usually associated with music and images, respectively.
2 Though you’ll know how to wrangle it shortly!
3 Actually, you won’t need that much luck—we’ll look at how to do this in “Wrangling PDFs with Python”.
4 In computer science, the terms data and information are applied in exactly the opposite way: data is the raw facts collected about the world, and information is the meaningful end product of organizing and structuring it. In recent years, however, as talk about “big data” has dominated many fields, the interpretation I use here has become more common, so I’ll stick with it throughout this book for clarity.
5 From the US Bureau of Labor Statistics.
6 “Multiple Jobholders” by Stéphane Auray, David L. Fuller, and Guillaume Vandenbroucke, posted December 21, 2018, https://research.stlouisfed.org/publications/economic-synopses/2018/12/21/multiple-jobholders.
7 See “New Recommendations on Improving Data on Contingent and Alternative Work Arrangements,” https://blogs.bls.gov/blog/tag/contingent-workers; “The Value of Flexible Work: Evidence from Uber Drivers” by M. Keith Chen et al., Nber Working Paper Series No. 23296, https://nber.org/system/files/working_papers/w23296/w23296.pdf.
8 You can also find instructions for this on the FRED website.
9 You can also download copies of these files directly from my Google Drive.
10 As of this writing, LibreOffice can handle the same number of rows as Microsoft Excel (220), but far fewer columns. While Google Sheets can handle more columns than Excel, it can only handle about 40,000 rows.
11 As of this writing, all of these libraries are already available and ready to use in Google Colab.
12 For more about get_rows(), see the xlrd documentation.
13 See the xlrd documentation for more on this issue.
14 From Stephen John Machin and Chris Withers, “Dates in Excel Spreadsheets”.
15 If you open the output files from the three preceding code examples in a text editor, you’ll notice that the open source .ods format is the simplest and cleanest.
16 As in, for example, Pennsylvania or Colorado.
17 See Vassili van der Mersch’s post, “Twitter’s 10 Year Struggle with Developer Relations” from Nordic APIs.
18 Unlike Python code, XML documents do not have to be properly indented in order to work, though it certainly makes them more readable!
19 Fun fact: the second x in the .xlsx format actually refers to XML!
20 Again, we’ll walk through using APIs like this one step by step in Chapter 5, but using this document lets us see how different data formats influence our interactions with the data.
21 If a stylesheet has been applied, as in the case of the BBC feed we used in Example 4-13, you can context+click the page and select “View Source” to see the “raw” XML.
22 See Adobe’s About PDF page for more information.
23 George Nagy, “Disruptive Developments in Document Recognition,” Pattern Recognition Letters 79 (2016): 106–112, https://doi.org/10.1016/j.patrec.2015.11.024. Available at https://ecse.rpi.edu/~nagy/PDF_chrono/2016_PRL_Disruptive_asPublished.pdf.
Get Practical Python Data Wrangling and Data Quality now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

