Chapter 4. Data Ingestion, Preprocessing, and Descriptive Statistics
You are most likely familiar with the phrase “garbage in, garbage out.” It captures well the notion that flawed, incorrect, or nonsensical data input will always produce faulty output. In the context of machine learning, it also emphasizes the fact that the attention we devote to ingesting, preprocessing, and statistically understanding our data (exploring and preparing it) will have an effect on the success of the overall process. Faulty data ingestion has a direct impact on the quality of the data, and so does faulty preprocessing. To get a feel for the data in hand, and its correctness, we leverage descriptive statistics; this is a vital part of the process as it helps us verify that the data we are using is of good quality. Data scientists, machine learning engineers, and data engineers often spend significant time working on, researching, and improving these crucial steps, and I will walk you through them in this chapter.
Before we start, let’s understand the flow. Let’s assume that at the beginning, our data resides on disk, in a database, or in a cloud data lake. Here are the steps we will follow to get an understanding of our data:
-
Ingestion. We begin by moving the data in its current form into a DataFrame instance. This is also called deserialization of the data. More accurately, in Spark, in this step we define a plan for how to deserialize the data, transforming it into a DataFrame. This step often provides us with a basic schema inferred from the existing data.
-
Preprocessing. This involves marshaling the data to fit into our desired schema. If we load the data as strings and we need it as floats, we will cast the data type and tweak the values as needed to fit the desired schema. This can be a complex and error-prone process, especially when synchronizing data from multiple sources at a multi-terabyte scale, and requires planning ahead.
-
Qualifying. This step consists of using descriptive statistics to understand the data and how to work with it.
Steps 2 and 3 can overlap, as we may decide to do more preprocessing on the data depending on the statistics calculated in step 3.
Now that you have a general idea of what the steps are, let’s dig a bit more deeply into each of them.
Data Ingestion with Spark
Apache Spark is generic enough to allow us to extend its API and develop dedicated connectors to any type of store for ingesting (and persisting/sinking/saving) data using the connector mechanism. Out of the box, it supports various file formats such as Parquet, CSV, binary files, JSON, ORC, image files, and more.
Spark also enables us to work with batch and streaming data. The Spark batch API is for processing offline data residing in a file store or database. With batch data, the dataset size is fixed and does not change, and we don’t get any fresh data to process. For processing streaming data, Spark has an older API called DStream or simply Streaming, and a newer, improved API called Structured Streaming. Structured Streaming provides an API for distributed continuous processing of structured data streams. It allows you to process multiple data records at a time, dividing the input stream into microbatches. Keep in mind that if your data is not structured, or if the format varies, you will need to use the legacy DStream API instead, or build a solution to automate schema changes without failures.
In this chapter, we will focus on batch processing with offline, cold data. Building machine learning models using cold data is the most common approach across varied use cases such as video production, financial modeling, drug discovery, genomic research, recommendation engines, and more. We will look at working with streaming data in Chapter 10, where we discuss serving models with both kinds of data sources.
Specifying batch reading with a defined data format is done using the format function, for example:
df=spark.read.format("image")
The class that enables this is the DataFrameReader class. You can configure it through its options API to define how to load the data and infer the schema if the file format doesn’t provide it already, or extract the metadata if it does.
Different file formats may either have a schema or not, depending on whether the data is structured, semi-structured, or unstructured, and, of course, the format’s implementation itself. For example, JSON format is considered semi-structured, and out of the box it doesn’t maintain metadata about the rows, columns, and features. So with JSON, the schema is inferred.
On the other hand, structured data formats such as Avro and Parquet have a metadata section that describes the data schema. This enables the schema to be extracted.
Working with Images
An image file can store data in an uncompressed, compressed, or vector format. For example, JPEG is a compressed format, and TIFF is an uncompressed format.
We save digital data in these formats in order to easily convert them for a computer display or a printer. This is the result of rasterization. Rasterization’s main task is converting the image data into a grid of pixels, where each pixel has a number of bits that define its color and transparency. Rasterizing an image file for a specific device takes into account the number of bits per pixel (the color depth) that the device is designed to handle. When we work with images, we need to attend to the file format and understand if it’s compressed or uncompressed (more on that in “Image compression and Parquet”).
In this chapter, we will use a Kaggle image dataset named Caltech 256 that contains image files with the JPEG compression format. Our first step will be to load them into a Spark DataFrame instance. For this, we can choose between two format options: image or binary.
Note
When your program processes the DataFrame load operation, the Spark engine does not immediately load the images into memory. As described in Chapter 2, Spark uses lazy evaluation, which means that instead of actually loading them, it creates a plan of how to load them if and when that becomes necessary. The plan contains information about the actual data, such as table fields/columns, format, file addresses, etc.
Image format
Spark MLlib has a dedicated image data source that enables us to load images from a directory into a DataFrame, which uses OpenCV types to read and process the image data. In this section, you will learn more about it.
OpenCV is a C/C++-based tool for computer vision workloads. MLlib functionality allows you to convert compressed images (.jpeg, .png, etc.) into the OpenCV data format. When loaded to a Spark DataFrame, each image is stored as a separate row.
The following are the supported uncompressed OpenCV types:
-
CV_8U
-
CV_8UC1
-
CV_8UC3
-
CV_8UC4
where 8 indicates the bit depth, U indicates unsigned, and Cx indicates the number of channels.
Warning
As of Spark 3.1.1, the limit on image size is 1 GB. Since Spark is open source, you can follow the image size support updates in its source: the line assert(imageSize < 1e9, "image is too large") in the definition of the decode function of the ImageSchema.scala object tells us that the limit is 1 GB.
To explore relatively small files, the image format Spark provides is a fantastic way to start working with images and actually see the rendered output. However, for the general workflow where there is no actual need to look at the images themselves during the process, I advise you to use the binary format, as it is more efficient and you will be able to process larger image files faster. Additionally, for larger images files (≥1 GB), binary format is the only way to process them. Whereas Spark’s default behavior is to partition data, images are not partitioned. With spatial objects such as images, we instead refer to tiling, or decomposing the image into a set of segments (tiles) with the desired shape. Tiling images can be part of the preprocessing of the data itself. Going forward, to align the language, I will refer to partitions even when discussing images or spatial objects.
Binary format
Spark began supporting a binary file data source with version 3.0.1 This enabled it to read binary files and convert them into a single record in a table. The record contains the raw content as BinaryType and a couple of metadata columns. Using binary format to read the data produces a DataFrame with the following columns:
-
path: StringType -
modificationTime: TimestampType -
length: LongType -
content: BinaryType
We’ll use this format to load the Caltech 256 data, as shown in the following code snippet:
from pyspark.sql.types import BinaryType
spark.sql("set spark.sql.files.ignoreCorruptFiles=true")
df = spark.read.format("binaryFile")
.option("pathGlobFilter", "*.jpg")
.option("recursiveFileLookup", "true")
.load(file_path)
As discussed in Chapter 2, the data within our dataset is in a nested folder hierarchy. recursiveFileLookup enables us to read the nested folders, while the pathGlobFilter option allows us to filter the files and read only the ones with a .jpg extension.
Again, note that this code does not actually load the data into the executors for computing. As discussed previously, because of Spark’s lazy evaluation mechanism, the execution will not start until an action is triggered—the driver accumulates the various transformation requests and queries in a DAG, optimizes them, and only acts when there is a specific request for an action. This allows us to save on computation costs, optimize queries, and increase the overall manageability of our code.
Working with Tabular Data
Since Spark provides out-of-the-box connectors to various file format types, it makes working with tabular data pretty straightforward. For example, in the book’s GitHub repository, under datasets you will find the CO2 Emission by Vehicles dataset, where the data is in CSV format. With the connector function .format("csv"), or directly with .csv(file_path), we can easily load this into a DataFrame instance.
Do pay attention to the schema, though—even with the InferSchema option, Spark tends to define columns in CSV files as containing strings even when they contain integers, Boolean values, etc. Hence, at the beginning, our main job is to check and correct the columns’ data types. For example, if a column in the input CSV file contains JSON strings, you’ll need to write dedicated code to handle this JSON.
Notice that each Spark data source connector has unique properties that provide you with a set of options for dealing with corrupted data. For example, you can control the column-pruning behavior by setting spark.sql.csv.parser.columnPruning.enabled to False if you don’t wish to prune columns with a corrupted format or data, or use True for the opposite behavior. You can also leverage the mode parameter to make pruning more specific, with approaches such as PERMISSIVE to set the field to null, DROPMALFORMED to ignore the whole record, or FAILFAST to throw an exception upon processing a corrupted record. See the following code snippet for an example:
df = spark.read.option("mode","FAILFAST")
.option("delimiter","\t")
.csv(file_path)
After loading the data and deserializing it into a DataFrame, it is time to preprocess it. Before moving on, I advise saving your data in a format that has a typed schema with well-defined column names and types, such as Parquet or Avro.
Preprocessing Data
Preprocessing is the art of transforming the data into the desired state, be it a strongly typed schema or a specific data type required by the algorithm.
Preprocessing Versus Processing
Differentiating between preprocessing and processing can be difficult when you’re just getting started with machine learning. Preprocessing refers to all the work we do before validating the dataset itself. This work is done before we attempt to get a feel for the data using descriptive statistics or perform feature engineering, both of which fall under the umbrella of processing. Those procedures are interlocked (see Figure 4-1), and we will likely repeat them again and again until we get the data into the desired state. Spark provides us with all the tools we need for these tasks, either through the MLlib library or SQL APIs.
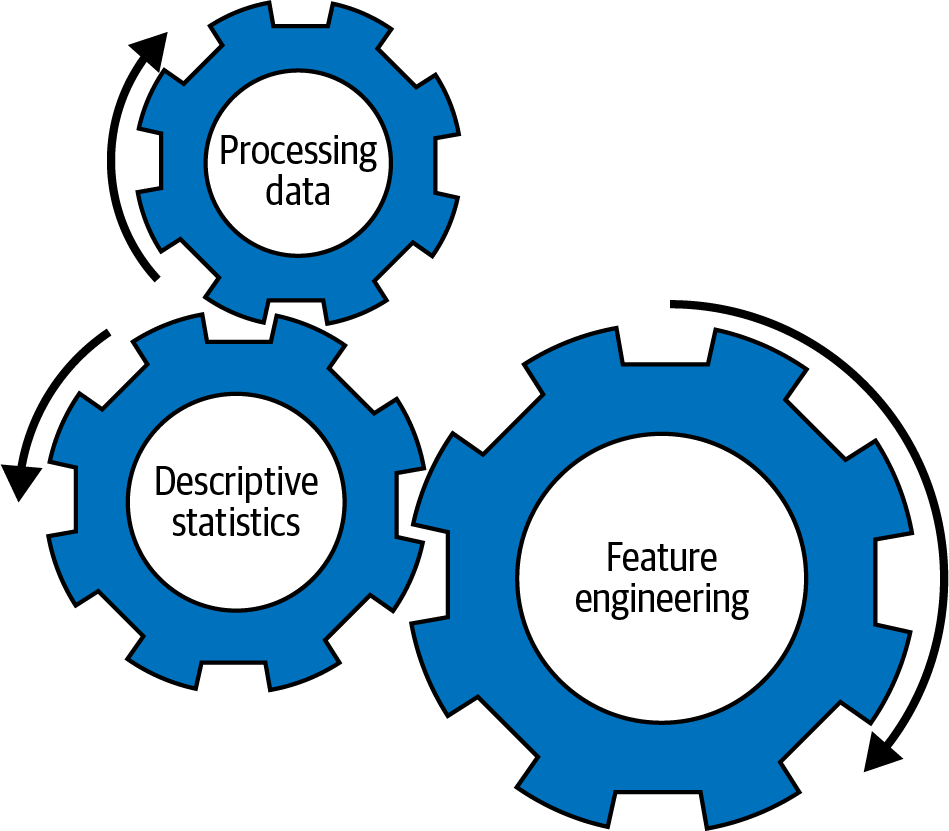
Figure 4-1. Interlocked procedures during machine learning
Why Preprocess the Data?
Preprocessing the data, or wrangling it into the desired schema, is a crucial step that must be completed before we can even start exploring the data, let alone engineering new features. The reason it’s so important is because machine learning algorithms often have dedicated input requirements, such as specific data structures and/or data types. In some academic research papers, you might find this process referred to as data marshaling.
To give you an idea of what kind of preprocessing you might need to perform on your data before passing it to an MLlib algorithm, let’s take a quick look at the high-level requirements of the different kinds of algorithms:
- Classification and/or regression algorithms
- For classification and regression, you will want to transform your data into one column of type
vector(dense or sparse) withdoubleorfloatvalues. This column is often namedfeatures, but you have the flexibility to set the input column name later. - Recommendation algorithms
- For recommendation, you will want to have a
userColcolumn withintegervalues representing the user IDs, anitemColcolumn withintegervalues representing the item IDs, and aratingColcolumn withdoubleorfloatvalues representing the items’ ratings by the users. - Unsupervised learning
- When dealing with data that calls for an unsupervised learning approach, you will often need one column of type
vectorto represent your features.
Data Structures
Depending on its structure, data often requires processing before it can be fully utilized. Data can be classified into three categories:
- Structured
- Structured data has a high degree of organization. It’s stored in a predefined schematic format, such as a comma-separated values (.csv) file or a table in a database. It is sometimes also referred to as tabular data.
- Semi-structured
- Semi-structured data has some degree of organization, but the structure is less rigid, and the schema is not fixed. There might be tags that separate elements and enforce hierarchies, like in a JSON file. Such data might require preprocessing before we can use it for machine learning algorithms.
- Unstructured
- Unstructured data has no defined organization and no specific format—think of .jpeg images, .mp4 video files, sound files, etc. This data often requires major preprocessing before we can use it to build models. Most of the data that we create today is unstructured data.
MLlib Data Types
MLlib has its own dedicated data types that it requires as input for machine learning algorithms. To work with Spark MLlib, you will need to transform your columns into one of these types—this is why preprocessing and transforming data are processes that you will perform in an interlocked manner. Under the hood, Spark uses the private objects VectorUDT and MatrixUDF, which abstract multiple types of local vectors (dense, sparse, labeled point) and matrices (both local and distributed). Those objects allow easy interaction with spark.sql.Dataset functionality. At a high level, the two types of objects are as follows:
- Vector
- A vector object represents a numeric vector. You can think of it as an array, just like in Python, only here the index is of type
integerand the value is of typedouble. - Matrix
- A matrix object represents a numeric matrix. It can be local to one machine or distributed across multiple machines. With the local version, indices are of type
integerand values are of typedouble. With the distributed version, indices are of typelongand values are of typedouble. All matrix types are represented by vectors.
Note
To simplify our work with Python tools, MLlib recognizes NumPy arrays and Python lists as dense vectors and SciPy’s csc_matrix with a single column as a sparse vector. This enables us to transition more easily from one tool to the other. Bear it in mind when working with multiple tools.
It’s good to understand how sparse and dense vectors are represented in MLlib, as you will encounter them in the documentation and your experiments. Spark decides which kind of vector to create for a given task behind the scenes.
Note
The third kind of vector is a labeled point; it represents features and labels of a data point and can be either dense or sparse. In MLlib, labeled points are used in supervised learning algorithms.
Let’s start with a dense vector. Here’s an example:
Row(features=DenseVector([1.2,543.5,0.0,0.0,0.0,1.0,0.0]))]
A DenseVector consists of a fixed-size array of values. It is considered less efficient than a SparseVector in terms of memory consumption because it explicitly creates memory space for the specified vector size, including any empty/default values. In our example, there are seven values in the DenseVector, but four of them are empty (0.0 is the default value, so these are considered empty values).
A SparseVector is an optimization of a DenseVector that has empty/default values. Let’s take a look at how our example DenseVector would look translated into a SparseVector:
Row(features=SparseVector(7,{0:1.2,1:543.5,5:1.0}))
The first number represents the size of the vector (7), and the map ({...}) represents the indices and their values. In this vector, there are only three values that need to be stored: the value at index 0 is 1.2, the value at index 2 is 543.5, and the value at index 5 is 1. The values at the other indices do not need to be stored, because they are all the default value.
Let’s take a look at a bigger vector example:
[Row(features=SparseVector(50,{48:9.9,49:6.7}))]
In this case, the vector size is 50, and we have only two values to store: 9.9 for index 48 and 6.7 for index 49.
A SparseVector can also look like this:
(50,[48,49],[9.9,6.7])
where the first number (here, 50) represents the size, the first array ([48,49]) represents the indices whose values are stored in the SparseVector, and the second array ([9.9,6.7]) represents the values at those indices. So in this example, the value at index 48 is 9.9, and the value at index 49 is 6.7. The rest of the vector indices all have a value of 0.0.
Why do we need both kinds of vectors? In machine learning, some algorithms, such as the naive Bayes classifier, work better on dense vector features and consequently might perform poorly given sparse vector features.
What can you do if your machine learning algorithm doesn’t work well with the kinds of features you have? First of all, the process described next will help you get a feel for your data and adapt it to fit your machine learning goal. If necessary, you can try collecting more data. You can also choose algorithms that perform better on sparse vectors. After all, this is part of the process of building machine learning models!
Tip
Make sure to bookmark and use the MLlib documentation. There is a community project dedicated to improving the Spark docs, which evolve and get better every day.
Preprocessing with MLlib Transformers
Transformers are part of the Apache Spark MLlib library named pyspark.ml.feature. In addition to transformers, it also provides extractors and selectors. Many of them are based on machine learning algorithms and statistical or mathematical computations. For preprocessing, we will leverage the transformers API, but you might find the other APIs helpful as well in different situations.
Transformers in Spark are algorithms or functions that take a DataFrame as an input and output a new DataFrame with the desired columns. In other words, you can think of them as translating a given input into a corresponding output. Transformers enable us to scale, convert, or modify existing columns. We can divide them loosely into the following categories: text data transformers, categorical feature transformers, continuous numerical transformers, and others.
The tables in the following sections will guide you on when to use each transformer. You should be aware that given the statistical nature of the transformers, some APIs may take longer to finish than others.
Working with text data
Text data typically consists of documents that represent words, sentences, or any form of free-flowing text. This is inherently unstructured data, which is often noisy. Noisy data in machine learning is irrelevant or meaningless data that might significantly impact the model’s performance. Examples include stopwords such as a, the, is, and are. In MLlib, you’ll find a dedicated function just for extracting stopwords, and so much more! MLlib provides a rich set of functionality for manipulating text data input.
With text, we want to ingest it and transform it into a format that can be easily fed into a machine learning algorithm. Most of the algorithms in MLlib expect structured data as input, in a tabular format with rows, columns, etc. On top of that, for efficiency in terms of memory consumption, we would typically hash the strings, since string values take up more space than integer, floating-point, or Boolean values. Before adding text data to your machine learning project, you first need to clean it up by using one of the text data transformers. To learn about the common APIs and their usage, take a look at Table 4-1.
| API | Usage |
|---|---|
Tokenizer |
Maps a column of text into a list of words by splitting on whitespace. It is based on the regular expression (regex) \\s, which matches a single whitespace character. Under the hood, the Tokenizer API uses the java.lang.String.split function. |
RegexTokenizer |
Splits the text based on the input regex (the default is \\s+, which matches one or more whitespace characters. It is typically used to split on whitespace and/or commas and other supported delimiters. RegexTokenizer is more computation-heavy than Tokenizer because it uses the scala.util.matching regex function. The supplied regex should conform to Java regular expression syntax. |
HashingTF |
Takes an array of strings and generates hashes from them. In many free-text scenarios, you will need to run the Tokenizer function first. This is one of the most widely used transformers. |
NGram |
Extracts a sequence of n tokens given an integer n. The input column can only be an array of strings. To convert a text string into an array of strings, use the Tokenizer function first. |
StopWordsRemover |
Takes a sequence of text and drops the default stopwords. You can specify languages and case sensitivity and provide your own list of stopwords. |
Before we proceed, let’s generate a synthetic dataset to use in the examples that follow:
sentence_data_frame=spark.createDataFrame([(0,"Hi I think pyspark is cool ","happy"),(1,"All I want is a pyspark cluster","indifferent"),(2,"I finally understand how ML works","fulfilled"),(3,"Yet another sentence about pyspark and ML","indifferent"),(4,"Why didn’t I know about mllib before","sad"),(5,"Yes, I can","happy")],["id","sentence","sentiment"])
Our dataset has three columns: id, of type int, and sentence and sentiment, of type string.
The transformation includes the following steps:
-
Free text → list of words
-
List of words → list of meaningful words
-
Select meaningful values
Ready? Set? Transform! Our first step is to transform the free text into a list of words. For that, we can use either the Tokenizer or the RegexTokenizer API, as shown here:
frompyspark.ml.featureimportTokenizertokenizer=Tokenizer(inputCol="sentence",outputCol="words")tokenized=tokenizer.transform(sentence_data_frame)
This tells Tokenizer to take the sentence column as an input and generate a new DataFrame, adding an output column named words. Notice that we used the transform function—transformers always have this function. Figure 4-2 shows our new DataFrame with the added words column.

Figure 4-2. New DataFrame with words column
The next step is to remove stopwords, or words that are unlikely to provide much value in our machine learning process. For that, we will use StopWordsRemover:
frompyspark.ml.featureimportStopWordsRemoverremover=StopWordsRemover(inputCol="words",outputCol="meaningful_words")meaningful_data_frame=remover.transform(tokenized)# I use the show function here for educational purposes only; with a large# dataset, you should avoid it.meaningful_data_frame.select("words","meaningful_words").show(5,truncate=False)
Example 4-1 shows the DataFrame with the new meaningful_words column.
Example 4-1. New DataFrame with meaningful_words column
+-------------------------------------------------+-------------------------------------+ |words |meaningful_words | +-------------------------------------------------+-------------------------------------+ |[hi, i, think, pyspark, is, cool] |[hi, think, pyspark, cool] | |[all, i, want, is, a, pyspark, cluster] |[want, pyspark, cluster] | |[i, finally, understand, how, ml, works] |[finally, understand, ml, works] | |[yet, another, sentence, about, pyspark, and, ml]|[yet, another, sentence, pyspark, ml]| |[why, didn't, i, know, about, ml lib, before] |[know, mllib] | |[yes,, i, can] |[yes,] | +-------------------------------------------------+-------------------------------------+
From nominal categorical features to indices
One of the strategies we can use to speed up the machine learning process is to turn discrete categorical values presented in string format into a numerical form using indices. The values can be discrete or continuous, depending on the machine learning models we plan to use. Table 4-2 lists the most common APIs and describes their usage.
| API | Usage |
|---|---|
StringIndexer |
Encodes string columns into indices, where the first one (starting at index 0) is the most frequent value in the column, and so on. Used for faster training with supervised data where the columns are the categories/labels. |
IndexToString |
The opposite of StringIndexer: maps a column of label indices back to a column containing the original labels as strings. Often used to retrieve the label categories after the training process. |
OneHotEncoder |
Maps a column of categorical features represented as label indices into a column of binary vectors, with at most one 1 value per row indicating the category. This allows machine learning algorithms that expect continuous features, such as logistic regression, to use categorical features by mapping them into continuous features. |
VectorIndexer |
Similar to StringIndexer; takes a vector column as input and converts it into category indices. |
The DataFrame we generated includes a column representing the sentiment of the text. Our sentiment categories are happy, fulfilled, sad, and indifferent. Let’s turn them into indices using StringIndexer:
frompyspark.ml.featureimportStringIndexerindexer=StringIndexer(inputCol="sentiment",outputCol="categoryIndex")indexed=indexer.fit(meaningful_data_frame).transform(meaningful_data_frame)indexed.show(5)
In this code snippet, we create a new StringIndexer instance that takes the sentiment column as an input and creates a new DataFrame with a categoryIndex column, of type double. We call the fit function first, providing it with the name of our DataFrame. This step is necessary for training the indexer before the transformation: it builds a map between indices and categories by scanning the sentiment column. This function is performed by another preprocessing tool called an estimator, which we’ll look at in more detail in Chapter 6. After fitting the estimator, we call transform to calculate the new indices. Example 4-2 shows the DataFrame with the new categoryIndex column.
Example 4-2. DataFrame with categoryIndex column
+---+--------------------+-----------+--------------------+--------------------+-------------+ | id| sentence| sentiment| words| meaningful_words|categoryIndex| +---+--------------------+-----------+--------------------+--------------------+-------------+ | O|Hi I think pyspar...| happy|[hi, i, think, py...|[i, think, pyspa... | 0.0| | 1|All I want is a p...|indifferent|[all, i, want, is...|[want, pyspark, c...| 1.0| | 2|I finally underst...| fulfilled|[i, finally, unde...|[finally, underst...| 2.0| | 3|Yet another sente...|indifferent|[yet, another, se...|[yet, another, se...| 1.0| | 4|Why didn't I know...| sad|[why, didn't, i, ...| [know, mllib]| 3.0| | 5| Yes, I can| happy| [yes,, i, can]| [yes,]| 0.0| +---+--------------------+-----------+--------------------+--------------------+-------------+
Structuring continuous numerical data
In some cases, we may have continuous numeric data that we want to structure. We do this by providing a threshold, or multiple thresholds, for taking an action or deciding on a classification.
Note
Continuous numeric values are often represented in a vector, with common data types being integer, float, and double.
For example, when we have scores for specific sentiments, as shown in Example 4-3, we can take an action when a given score falls into a defined range. Think about a customer satisfaction system—we would like our machine learning model to recommend an action that is based on customer sentiment scores. Let’s say our biggest customer has a sad score of 0.75 and our threshold for calling the customer to discuss how we can improve their experience is a sad score above 0.7. In this instance, we would want to call the customer. The thresholds themselves can be defined manually or by using machine learning algorithms or plain statistics. Going forward, let’s assume we have a DataFrame with a dedicated score for every sentiment. That score is a continuous number in the range [0,1] specifying the relevancy of the sentiment category. The business goal we want to achieve will determine the thresholds to use and the structure to give the data for future recommendations.
Example 4-3. DataFrame with sentiment score for each category
+-----------+-----+-----------+---------+----+ |sentence_id|happy|indifferent|fulfilled| sad| +-----------+-----+-----------+---------+----+ | 0| 0.01| 0.43| 0.3| 0.5| | 1|0.097| 0.21| 0.2| 0.9| | 2| 0.4| 0.329| 0.97| 0.4| | 3| 0.7| 0.4| 0.3|0.87| | 4| 0.34| 0.4| 0.3|0.78| | 5| 0.1| 0.3| 0.31|0.29| +-----------+-----+-----------+---------+----+
Take into consideration the type of data you are working with and cast it when necessary. You can leverage the Spark SQL API for this, as shown here:
cast_data_frame=sentiment_data_frame.selectExpr("cast(happy as double)")
The following are some common strategies for handling continuous numerical data:
- Fixed bucketing/binning
- This is done manually by either binarizing the data by providing a specific threshold or providing a range of buckets. This process is similar to what we discussed earlier with regard to structuring continuous data.
- Adaptive bucketing/binning
- The overall data may be skewed, with some values occurring frequently while others are rare. This might make it hard to manually specify a range for each bucket. Adaptive bucketing is a more advanced technique where the transformer calculates the distribution of the data and sets the bucket sizes so that each one contains approximately the same number of values.
Table 4-3 lists the most commonly used continuous numerical transformers available in MLlib. Remember to pick the one that fits your project based on your needs.
| API | Usage |
|---|---|
Binarizer |
Turns a numerical feature into a binary feature, given a threshold. For example, 5.1 with threshold 0.7 would turn into 1, and 0.6 would turn into 0. |
Bucketizer |
Takes a column of continuous numerical values and transforms it into a column of buckets, where each bucket represents a part of the range of values—for example, 0 to 1, 1 to 2, and so on. |
MaxAbsScaler |
Takes a vector of float values and divides each value by the maximum absolute value in the input columns. |
MinMaxScaler |
Scales the data to the desired min and max values, where the default range is [0,1]. |
Normalizer |
Converts a vector of double values into normalized values that are nonnegative real numbers between 0 and 1. The default p-norm is 2, which implements the Euclidean norm for calculating a distance and reducing the float range to [0,1]. |
QuantileDiscretizer |
Takes a column of continuous numerical values and transforms it into a column with binned categorical values, with the input maximum number of bins optionally determining the approximate quantile values. |
RobustScaler |
Similar to StandardScaler; takes a vector of float values and produces a vector of scaled features given the input quantile range. |
StandardScaler |
Estimator that takes a vector of float values and aims to center the data given the input standard deviation and mean value. |
Additional transformers
MLlib offers many additional transformers that use statistics or abstract other Spark functionality. Table 4-4 lists some of these and describes their usage. Bear in mind that more are added regularly, with code examples available in the examples/src/main/python/ml/ directory of the Apache Spark GitHub repository.
| API | Usage |
|---|---|
DCT |
Implements a discrete cosine transform, taking a vector of data points in the time domain and translating them to the frequency domain. Used in signal processing and data compression (e.g., with images, audio, radio, and digital television). |
ElementwiseProduct |
Takes a column with vectors of data and a transforming vector of the same size and outputs a multiplication of them that is associative, distributive, and commutative (based on the Hadamard product). This is used to scale the existing vectors. |
Imputer |
Takes a column of numeric type and completes missing values in the dataset using the column mean or median value. Useful when using estimators that can’t handle missing values. |
Interaction |
Takes one distinct vector or double-valued column and outputs a vector column containing the products of all the possible combinations of values. |
PCA |
Implements principal component analysis, turning a vector of potentially correlated values into non-correlated ones by outputting the most important components of the data (the principal components). This is useful in predictive models and dimensionality reduction, with a potential cost of interpretability. |
PolynomialExpansion |
Takes a vector of features and expands it into an n-degree polynomial space. A value of 1 means no expansion. |
SQLTransformer |
Takes an SQL statement (any SELECT clause that Spark supports) and transforms the input according to the statement. |
VectorAssembler |
Takes a list of vector columns and concatenates them into one column in the dataset. This is useful for various estimators that take only one column. |
Preprocessing Image Data
Image data is common in machine learning applications, and it also requires preprocessing to move forward in the machine learning workflow. But images are different from the kinds of data we’ve seen before, and they require a different kind of procedure. There may be more or fewer steps involved, depending on the actual data, but the most common path consists of these three actions:
-
Extract labels
-
Transform labels to indices
-
Extract image size
Let’s walk through these steps using our example dataset to see what they involve.
Extracting labels
Our images dataset has a nested structure where the directory name indicates the classification of the image. Hence, each image’s path on the filesystem contains its label. We need to extract this in order to use it later on. Most raw image datasets follow this pattern, and this is an essential part of the preprocessing we will do with our images. After loading the images as BinaryType, we get a table with a column called path of type String. This contains our labels. Now, it’s time to leverage string manipulation to extract that data. Let’s take a look at one example path: .../256_ObjectCategories/198.spider/198_0089.jpg.
The label in this case is actually an index and a name: 198.spider. This is the part we need to extract from the string. Fortunately, PySpark SQL functions provide us with the regexp_extract API that enables us to easily manipulate strings according to our needs.
Let’s define a function that will take a path_col and use this API to extract the label, with the regex "256_ObjectCategories/([^/]+)":
frompyspark.sql.functionsimportcol,regexp_extractdefextract_label(path_col):"""Extract label from file path using built-in SQL function"""returnregexp_extract(path_col,"256_ObjectCategories/([^/]+)",1)
We can now create a new DataFrame with the labels by calling this function from a Spark SQL query:
images_with_label=df_result.select(col("path"),extract_label(col("path")).alias("label"),col("content"))
Our images_with_label DataFrame consists of three columns: two string columns called path and label and a binary column called content.
Now that we have our labels, it’s time to transform them into indices.
Transforming labels to indices
As mentioned previously, our label column is a string column. This can pose a challenge for machine learning models, as strings tend to be heavy on memory usage. Ideally, every string in our table should be transformed into a more efficient representation before being ingested into a machine learning algorithm, unless it is a true necessity not to. Since our labels are of the format {index.name}, we have three options:
-
Extract the index from the string itself, leveraging string manipulation.
-
Provide a new index using Spark’s
StringIndexer, as discussed in “From nominal categorical features to indices”. -
Use Python to define an index (in the Caltech 256 dataset, there are only 257 indices, in the range
[1,257]).
In our case, the first option is the cleanest way to handle this. This approach will allow us to avoid maintaining a mapping between the indices in the original files and the indices in the dataset.
Extracting image size
The final step is to extract the image size. We do this as part of our preprocessing because we know for sure that our dataset contains images of different sizes, but it’s often a useful operation to get a feel for the data and provide us with information to help us decide on an algorithm. Some machine learning algorithms will require us to have a unified size for images, and knowing in advance what image sizes we’re working with can help us make better optimization decisions.
Since Spark doesn’t yet provide this functionality out of the box, we’ll use Pillow (aka PIL), a friendly Python library for working with images. To efficiently extract the width and height of all of our images, we will define a pandas user-defined function (UDF) that can run in a distributed fashion on our Spark executors. pandas UDFs, defined using pandas_udf as a decorator, are optimized with Apache Arrow and are faster for grouped operations (e.g., when applied after a groupBy).
Grouping allows pandas to perform vectorized operations. For these kinds of use cases, a pandas UDF on Spark will be more efficient. For simple operations like a*b, a Spark UDF will suffice and will be faster because it has less overhead.
Our UDF will take a series of rows and operate on them in parallel, making it much faster than the traditional one-row-at-a-time approach:
frompyspark.sql.functionsimportcol,pandas_udffromPILimportImageimportpandasaspd@pandas_udf("width: int, height: int")defextract_size_udf(content_series):sizes=content_series.apply(extract_size)returnpd.DataFrame(list(sizes))
Now that we have the function, we can pass it to Spark’s select function to extract the image size information:
images_df=images_with_label.select(col("path"),col("label"),extract_size_udf(col("content")).alias("size"),col("content"))
The image size data will be extracted into a new DataFrame with a size column of type struct containing the width and height:
size:structwidth:integerheight:integer
Warning
Note that using the extract_size_udf function will transfer all of the images from the JVM (Spark uses Scala under the hood) to the Python runtime using Arrow, compute the sizes, and then transfer the sizes back to the JVM. For efficiency when working with large datasets, especially if you are not using grouping, it might be worth implementing the extraction of the sizes at the JVM/Scala level instead. Keep considerations like this in mind when implementing data preprocessing for the various stages of machine learning.
Save the Data and Avoid the Small Files Problem
When you’ve completed all of your preprocessing, it can be a good idea to save the data to cold or hot storage before continuing to the next step. This is sometimes referred to as a checkpoint, or a point in time where we save a version of the data we are happy with. One reason to save the data is to enable fast recovery: if our Spark cluster breaks completely, instead of needing to recalculate everything from scratch, we can recover from the last checkpoint. The second reason is to facilitate collaboration. If your preprocessed data is persisted to storage and available for your colleagues to use, they can leverage it to develop their own flows. This is especially useful when working with large datasets and on tasks requiring extensive computation resources and time. Spark provides us with functionality to both ingest and save data in numerous formats. Note that if you decide to save the data for collaboration purposes, it’s important to document all the steps that you took: the preprocessing you carried out, the code you used to implement it, the current use cases, any tuning you performed, and any external resources you created, like stopword lists.
Avoiding small files
A small file is any file that is significantly smaller than the storage block size. Yes, there is a minimum block size, even with object stores such as Amazon S3, Azure Blob, etc.! Having files that are significantly smaller than the block size can result in wasted space on the disk, since the storage will use the whole block to save that file, no matter how small it is. This is an overhead that we should avoid. On top of that, storage is optimized to support fast reading and writing by block size. But don’t worry—Spark API to the rescue! We can easily avoid wasting precious space and paying a high price for storing small files using Spark’s repartition or coalesce functions.
In our case, because we are operating on offline data without a specific requirement to finish computation on the millisecond, we have more flexibility in which one to choose. repartition creates entirely new partitions, shuffling the data over the network with the goal of distributing it evenly over the specified number of partitions (which can be higher or lower than the existing number). It therefore has a high cost up front, but down the road, Spark functionality will execute faster because the data will be distributed optimally—in fact, executing a repartition operation can be useful in any stage of the machine learning workflow when we notice that computation is relatively slow and want to speed it up. The coalesce function, on the other hand, first detects the existing partitions and then shuffles only the necessary data. It can only be used to reduce the number of partitions, not to add partitions, and is known to run faster than repartition as it minimizes the amount of data shuffled over the network. In some cases, coalesce might not shuffle at all and will default to batch local partitions, which makes it super efficient for reduce functionality.
Since we want to be in control of the exact number of partitions and we don’t require extremely fast execution, in our case it’s okay to use the slower repartition function, as shown here:
output_df=output_df.repartition(NUM_EXECUTERS)
Bear in mind that if time, efficiency, and minimizing network load are of the essence, you should opt for the coalesce function instead.
Image compression and Parquet
Suppose we want to save our image dataset in Parquet format (if you’re not familiar with it, Parquet is an open source, column-oriented data file format designed for efficient storage and retrieval). When saving to this format, by default Spark uses a compression codec named Snappy. However, since images are already compressed (e.g., with JPEG, PNG, etc.), it wouldn’t make sense to compress them again. How can we avoid this?
We save the existing configured compression codec in a string instance, configure Spark to write to Parquet with the uncompressed codec, save the data in Parquet format, and assign the codec instance back to the Spark configuration for future work. The following code snippet demonstrates:
# Image data is already compressed, so we turn off Parquet compressioncompression=spark.conf.get("spark.sql.parquet.compression.codec")spark.conf.set("spark.sql.parquet.compression.codec","uncompressed")# Save the data stored in binary format as Parquetoutput_df.write.mode("overwrite").parquet(save_path)spark.conf.set("spark.sql.parquet.compression.codec",compression)
Descriptive Statistics: Getting a Feel for the Data
Machine learning is not magic—you will need to understand your data in order to work with it efficiently and effectively. Getting a solid understanding of the data before you start training your algorithms will save you much time and effort down the road. Fortunately, MLlib provides a dedicated library named pyspark.ml.stat that contains all the functionality you need for extracting basic statistics out of the data.
Don’t worry if that sounds intimidating—you don’t need to fully understand statistics to use MLlib, although some level of familiarity will definitely help you in your machine learning journey. Understanding the data using statistics enables us to better decide on which machine learning algorithm to use, identify biases, and estimate the quality of the data—as mentioned previously, if you put garbage in, you get garbage out. Ingesting low-quality data into a machine learning algorithm will result in a low-performing model. As a result, this part is a must!
Having said that, as long as we build conscious assumptions about what the data looks like, what we can accept, and what we cannot, we can conduct much better experiments and have a better idea of what to remove, what to input, and what we can be lenient about. Take into consideration that those assumptions and any data cleansing operations we perform can have big consequences in production, especially if they are aggressive (like dropping all nulls in a large number of rows or imputing too many default values, which screws up the entropy completely). Watch out for mismatches in assumptions made during the exploratory stages about the data input, quality measurements, and what constitutes “bad” or low-quality data.
Tip
For deeper statistical analysis of a given dataset, many data scientists use the pandas library. As mentioned in Chapter 2, pandas is a Python analysis library for working with relatively small data that can fit in one machine’s memory (RAM). Its counterpart in the Apache Spark ecosystem is Koalas, which has evolved into a pandas API on Spark. There is not 100% feature parity between the original pandas and pandas on Spark, but this API expands Spark’s capabilities and takes them even further, so it’s worth checking out.
In this section, we will shift gears from straightforward processes and will focus on getting a feel for the data with the Spark MLlib functionality for computing statistics.
Calculating Statistics
Welcome to the Machine Learning Zoo Project!
For learning about MLlib’s statistics functions, we’ll use the Zoo Animal Classification dataset from the Kaggle repository. This dataset, created in 1990, consists of 101 examples of zoo animals described by 16 Boolean-valued attributes capturing various traits. The animals can be classified into seven types: Mammal, Bird, Reptile, Fish, Amphibian, Bug, and Invertebrate.
The first thing you need to do to get a feel for the data to better plan your machine learning journey is calculate the feature statistics. Knowing how the data itself is distributed will provide you with valuable insights to determine which algorithms to select, how to evaluate the model, and overall how much effort you need to invest in the project.
Descriptive Statistics with Spark Summarizer
A descriptive statistic is a summary statistic that quantitatively describes or summarizes features from a collection of information. MLlib provides us with a dedicated Summarizer object for computing statistical metrics from a specific column. This functionality is part of the MLlib LinearRegression algorithm for building the LinearRegressionSummary. When building the Summarizer, we need to specify the desired metrics. Table 4-5 lists the functionality available in the Spark API.
| Metric | Description |
|---|---|
mean |
Calculates the average value of a given numerical column |
sum |
Calculates the sum of the numerical column |
variance |
Calculates the variance of the column (how far the set of numbers in the column are spread out from its mean value, on average) |
std |
Calculates the standard deviation of the column (the square root of the variance value), to provide more weight to outliers in the column |
count |
Calculates the number of items/rows in the dataset |
numNonZeros |
Finds the number of nonzero values in the column |
max |
Finds the maximum value in the column |
min |
Finds the minimum value in the column |
normL1 |
Calculates the L1 norm (similarity between the numeric values) of the column |
normL2 |
Calculates the Euclidean norm of the column |
Note
The L1 and L2 (aka Euclidean) norms are tools for calculating the distance between numeric points in an N-dimensional space. They are commonly used as metrics to measure the similarity between data points in fields such as geometry, data mining, and deep learning.
This code snippet illustrates how to create a Summarizer instance with metrics:
frompyspark.ml.statimportSummarizersummarizer=Summarizer.metrics("mean","sum","variance","std")
Like the other MLlib functions, the Summarizer.metrics function expects a vector of numeric features as input. You can use MLlib’s VectorAssembler function to assemble the vector.
Although there are many features in the Zoo Animal Classification dataset, we are going to examine just the following columns:
-
feathers -
milk -
fins -
domestic
As discussed in “Data Ingestion with Spark”, we’ll load the data into a DataFrame instance named zoo_data_for_statistics.
In the next code sample, you can see how to build the vector. Notice how we set the output column name to features, as expected by the summarizer:
frompyspark.ml.featureimportVectorAssembler# set the output col to features as expected as input for the summarizervecAssembler=VectorAssembler(outputCol="features")# assemble only part of the columns for the examplevecAssembler.setInputCols(["feathers","milk","fins","domestic"])vector_df=vecAssembler.transform(zoo_data_for_statistics)
Our vector is leveraging Apache Spark’s Dataset functionality. A Dataset in Spark is a strongly typed collection of objects encapsulating the DataFrame. You can still call the DataFrame from a Dataset if needed, but the Dataset API enables you to access a specific column without the dedicated column functionality:
Vector_df.features
Now that we have a dedicated vector column and a summarizer, let’s extract some statistics. We can call the summarizer.summary function to plot all the metrics or to compute a specific metric, as shown in the following example:
# compute statistics for multiple metricsstatistics_df=vector_df.select(summarizer.summary(vector_df.features))# statistics_df will plot all the metricsstatistics_df.show(truncate=False)# compute statistics for single metric (here, std) without the restvector_df.select(Summarizer.std(vector_df.features)).show(truncate=False)
Example 4-4 shows the output of calling std on the vector of features.
Example 4-4. std of the features column
+-------------------------------------------------------------------------------+ |std(features) | +-------------------------------------------------------------------------------+ |[0.4004947435409863,0.4935223970962651,0.37601348195757744,0.33655211592363116]| +-------------------------------------------------------------------------------+
The standard deviation (STD) is an indicator of the variation in a set of values. A low STD indicates that the values tend to be close to the mean (also called the expected value) of the set, while a high STD indicates that the values are spread out over a wider range.
Since the features feathers, milk, fins, and domestic are inherently of type Boolean—milk can be 1 for true or 0 for false, and the same for fins and so on—calculating the STD doesn’t provide us with much insight—the result will always be a decimal number between 0 and 1. That misses the value of STD in calculating how “spread out” the data is. Instead, let’s try the sum function. This function will tell us how many animals in the dataset have feathers, milk, or fins or are domestic animals:
# compute statistics for single metric "sum" without the restvector_df.select(Summarizer.sum(vector_df.features)).show(truncate=False)
Take a look at the output of sum, shown in Example 4-5.
+---------------------+ |sum(features) | +---------------------+ |[20.0,41.0,17.0,13.0]| +---------------------+
This tells us that there are 20 animals with feathers (the vector’s first value), 41 animals that provide milk (the vector’s second value), 17 animals with fins (the third value), and 13 domestic animals (the final value). The sum function provides us with more insights about the data itself than the std function, due to the Boolean nature of the data. However, the more complicated/diverse the dataset is, the more looking at all of the various metrics will help.
Data Skewness
Skewness in statistics is a measure of the asymmetry of a probability distribution. Think of a bell curve where the data points are not distributed symmetrically on the left and right sides of the curve’s mean value. Assuming the dataset follows a normal distribution curve, skewness means it has a short tail on one end and a long tail on the other. The higher the skewness value is, the less evenly distributed the data is, and the more data points will fall on one side of the bell curve.
To measure skewness, or the asymmetry of the values around the mean, we need to extract the mean value and calculate the standard deviation. A statistical equation to accomplish this has already been implemented in Spark for us; check out the next code snippet to see how to take advantage of it:
from pyspark.sql.functions import skewness
df_with_skew = df.select(skewness("{column_name}"))
This code returns a new DataFrame with a dedicated column that measures the skewness of the column we requested. Spark also implements other statistical functions, such as kurtosis, which measures the tail of the data. Both are important when building a model based on the distribution of random variables and the assumption that the data follows a normal distribution; they can help you detect biases, data topology changes, and even data drift. We’ll discuss data drift in more detail in Chapter 10, when we look at monitoring machine learning models in production).
Correlation
A correlation between two features means that if feature A increases or decreases, feature B does the same (a positive correlation) or does the exact opposite (a negative correlation). Determining correlation therefore involves measuring the linear relationship between the two variables/features. Since a machine learning algorithm’s goal is to learn from data, perfectly correlated features are less likely to provide insights to improve model accuracy. This is why filtering them out can significantly improve our algorithm’s performance while maintaining the quality of the results. The test method of the ChiSquareTest class in MLlib is a statistical test that helps us assess categorical data and labels by running a Pearson correlation on all pairs and outputting a matrix with correlation scores.
Warning
Be mindful that correlation doesn’t necessarily imply causation. When the values of two variables change in a correlated way, there is no guarantee that the change in one variable causes the change in the other. It takes more effort to prove a causative relationship.
In this section, you will learn about Pearson and Spearman correlations in Spark MLlib.
Pearson correlation
When looking into correlation, we look for positive or negative associations. Pearson correlation measures the strength of linear association between two variables. It produces a coefficient r that indicates how far away the data points are from a descriptive line. The range of r is [–1,1], where:
-
r=1is a perfect positive correlation. Both variables act in the same way. -
r=–1is perfect negative/inverse correlation, which means that when one variable increases, the other decreases. -
r=0means no correlation.
Figure 4-3 shows some examples on a graph.
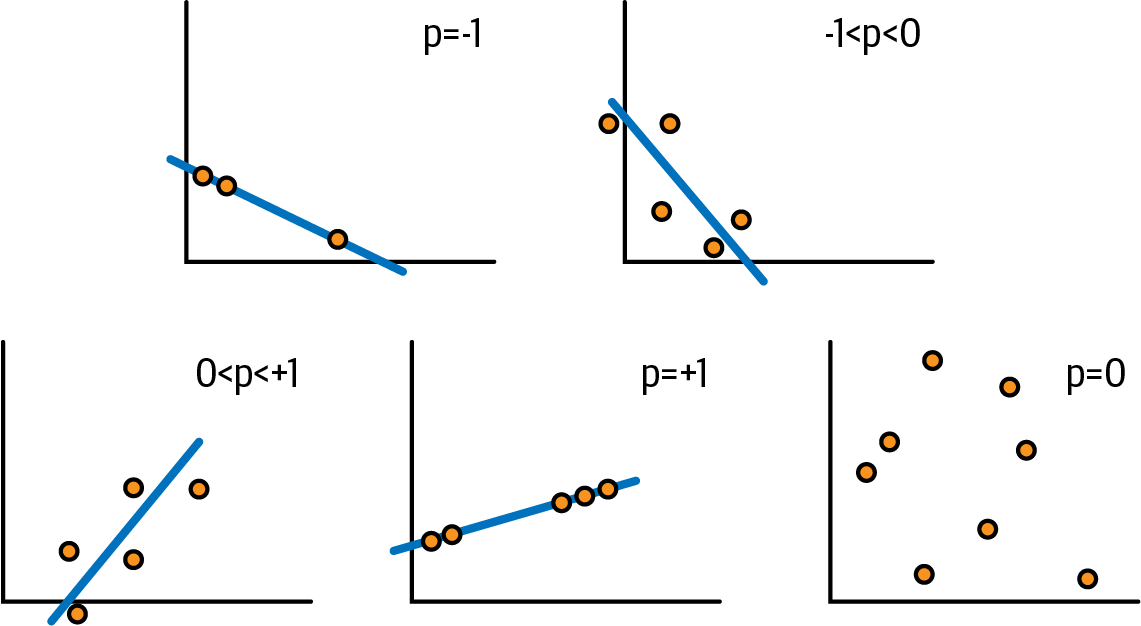
Figure 4-3. Pearson correlation examples on a graph
Pearson is the default correlation test with the MLlib Correlation object. Let’s take a look at some example code and its results:
frompyspark.ml.statimportCorrelation# compute r1 0 Pearson correlationr1=Correlation.corr(vector_df,"features").head()("Pearson correlation matrix:\n"+str(r1[0])+"\n")
The output is a row where the first value is a DenseMatrix, as shown in Example 4-6.
Example 4-6. Pearson correlation matrix
Pearson correlation matrix:
DenseMatrix([[ 1. , -0.41076061, -0.22354106, 0.03158624],
[-0.41076061, 1. , -0.15632771, 0.16392762],
[-0.22354106, -0.15632771, 1. , -0.09388671],
[ 0.03158624, 0.16392762, -0.09388671, 1. ]])
Each line represents the correlation of a feature with all the other features, in a pairwise way: for example, r1[0][0,1] represents the correlation of feathers with milk, which is a negative value (-0.41076061) that indicates a negative correlation between animals that produce milk and animals with feathers.
Table 4-6 shows what the correlation table looks like, to make this clearer.
feathers |
milk |
fins |
domestic |
|
|---|---|---|---|---|
feathers |
1 |
-.41076061 |
-0.22354106 |
0.03158624 |
milk |
-0.41076061 |
1 |
-0.15632771 |
0.16392762 |
fins |
-0.22354106 |
-0.15632771 |
1 |
-0.09388671 |
domestic |
0.03158624 |
0.16392762 |
-0.09388671 |
1 |
This table makes it easy to spot negative and positive correlations: for example, fins and milk have a negative correlation, while domestic and milk have a positive correlation.
Spearman correlation
Spearman correlation, also known as Spearman rank correlation, measures the strength and direction of the monotonic relationship between two variables. In contrast to Pearson, which measures the linear relationship, this is a curvilinear relationship, which means the association between the two variables changes as the values change (increase or decrease). Spearman correlation should be used when the data is discrete and the relationships between the data points are not necessarily linear, as shown in Figure 4-4, as well as when ranking is of interest. To decide which approach fits your data better, you need to understand the nature of the data itself: if it’s on an ordinal scale,2 use Spearman, and if it’s on an interval scale,3 use Pearson. To learn more about this, I recommend reading Practical Statistics for Data Scientists by Peter Bruce, Andrew Bruce, and Peter Gedeck (O’Reilly).
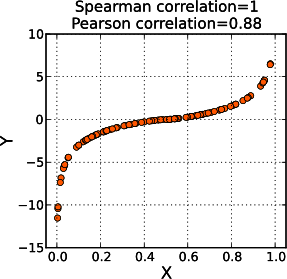
Figure 4-4. An example showing that Spearman correlation plots do not create a linear graph (image source: Wikipedia, CC BY-SA 3.0)
Notice that to use Spearman, you have to specify it (the default is Pearson):
frompyspark.ml.statimportCorrelation# compute r2 0 Spearman correlationr2=Correlation.corr(vector_df,"features","spearman").head()("Spearman correlation matrix:\n"+str(r2[0]))
As before, the output is a row where the first value is a DenseMatrix, and it follows the same rules and order described previously (see Example 4-7).
Example 4-7. Spearman correlation matrix
Spearman correlation matrix: DenseMatrix([[ 1. , -0.41076061, -0.22354106, 0.03158624], [-0.41076061, 1. , -0.15632771, 0.16392762], [-0.22354106, -0.15632771, 1. , -0.09388671], [ 0.03158624, 0.16392762, -0.09388671, 1. ]])
Tip
Spark has an automation for feature selectors based on correlation and hypothesis tests, such as chi-squared, ANOVA F-test, and F-value (the UnivariateFeatureSelector; see Table 5-2 in Chapter 5). To speed up the process, it is best to use existing, implemented tests instead of calculating every hypothesis by yourself. If after the feature selection process you identify an insufficient set of features, you should use hypothesis tests such as ChiSquareTest to evaluate whether you need to enrich your data or find a larger set of data. I’ve provided you with a code example in the book’s GitHub repository demonstrating how to do this. Statistical hypothesis tests have a null hypothesis (H0) and alternative hypothesis (H1), where:
- H0: The sample data follows the hypothesized distribution.
- H1: The sample data does not follow the hypothesized distribution.
The outcome of the test is the p-value, which demonstrates the likelihood of H0 being true.
Summary
In this chapter, we discussed three crucial steps in the machine learning workflow: ingesting data, preprocessing text and images, and gathering descriptive statistics. Data scientists and machine learning engineers typically spend a significant portion of their time on these tasks, and executing them mindfully sets us up for greater success and a better machine learning model that can meet the business goal in a much more profound way. As a rule of thumb, it’s best to collaborate with your peers to validate and engineer these steps to ensure the resulting insights and data can be reused in multiple experiments. The tools introduced in this chapter will accompany us again and again throughout the machine learning process. In the next chapter, we will dive into feature engineering and build upon the outcomes from this chapter.
1 The binary data source schema may change with new releases of Apache Spark or when using Spark in managed environments such as Databricks.
2 An ordinal scale has all its variables in a specific order, beyond just naming them.
3 An interval scale labels and orders its variables and specifies a defined, evenly spaced interval between them.
Get Scaling Machine Learning with Spark now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

