Preface
Programming is a force multiplier. We can write computer programs to free ourselves from tedious manual tasks and to accelerate research. Programming in any language will likely improve your productivity, but each language has different learning curves and tools that improve or impede the process of coding.
There is an adage in business that says you have three choices:
-
Fast
-
Good
-
Cheap
Pick any two.
When it comes to programming languages, Python hits a sweet spot in that itâs fast because itâs fairly easy to learn and to write a working prototype of an ideaâitâs pretty much always the first language Iâll use to write any program. I find Python to be cheap because my programs will usually run well enough on commodity hardware like my laptop or a tiny AWS instance. However, I would contend that itâs not necessarily easy to make good programs using Python because the language itself is fairly lax. For instance, it allows one to mix characters and numbers in operations that will crash the program.
This book has been written for the aspiring bioinformatics programmer who wants to learn about Pythonâs best practices and tools such as the following:
-
Since Python 3.6, you can add type hints to indicate, for instance, that a variable should be a type like a number or a list, and you can use the
mypytool to ensure the types are used correctly. -
Testing frameworks like
pytestcan exercise your code with both good and bad data to ensure that it reacts in some predictable way. -
Tools like
pylintandflake8can find potential errors and stylistic problems that would make your programs more difficult to understand. -
The
argparsemodule can document and validate the arguments to your programs. -
The Python ecosystem allows you to leverage hundreds of existing modules like Biopython to shorten programs and make them more reliable.
Using these tools practices individually will improve your programs, but combining them all will improve your code in compounding ways. This book is not a textbook on bioinformatics per se. The focus is on what Python offers that makes it suitable for writing scientific programs that are reproducible. That is, Iâll show you how to design and test programs that will always produce the same outputs given the same inputs. Bioinformatics is saturated with poorly written, undocumented programs, and my goal is to reverse this trend, one program at a time.
The criteria for program reproducibility include:
- Parameters
-
All program parameters can be set as runtime arguments. This means no hard-coded values which would require changing the source code to change the programâs behavior.
- Documentation
-
A program should respond to a
--helpargument by printing the parameters and usage. - Testing
-
You should be able to run a test suite that proves the code meets some specifications
You might expect that this would logically lead to programs that are perhaps correct, but alas, as Edsger Dijkstra famously said, âProgram testing can be used to show the presence of bugs, but never to show their absence!â
Most bioinformaticians are either scientists whoâve learned programming or programmers whoâve learned biology (or people like me who had to learn both). No matter how youâve come to the field of bioinformatics, I want to show you practical programming techniques that will help you write correct programs quickly. Iâll start with how to write programs that document and validate their arguments. Then Iâll show how to write and run tests to ensure the programs do what they purport.
For instance, the first chapter shows you how to report the tetranucleotide frequency from a string of DNA. Sounds pretty simple, right? Itâs a trivial idea, but Iâll take about 40 pages to show how to structure, document, and test this program. Iâll spend a lot of time on how to write and test several different versions of the program so that I can explore many aspects of Python data structures, syntax, modules, and tools.
Who Should Read This?
You should read this book if you care about the craft of programming, and if you want to learn how to write programs that produce documentation, validate their parameters, fail gracefully, and work reliably. Testing is a key skill both for understanding your code and for verifying its correctness. Iâll show you how to use the tests Iâve written as well as how to write tests for your programs.
To get the most out of this book, you should already have a solid understanding of Python.
I will build on the skills I taught in Tiny Python Projects (Manning, 2020), where I show how to use Python data structures like strings, lists, tuples, dictionaries, sets, and named tuples.
You need not be an expert in Python, but I definitely will push you to understand some advanced concepts I introduce in that book, such as types, regular expressions, and ideas about higher-order functions, along with testing and how to use tools like pylint, flake8, yapf, and pytest to check style, syntax, and correctness.
One notable difference is that I will consistently use type annotations for all code in this book and will use the mypy tool to ensure the correct use of types.
Programming Style: Why I Avoid OOP and Exceptions
I tend to avoid object-oriented programming (OOP). If you donât know what OOP means, thatâs OK. Python itself is an OO language, and almost every element from a string to a set is technically an object with internal state and methods. You will encounter enough objects to get a feel for what OOP means, but the programs I present will mostly avoid using objects to represent ideas.
That said, Chapter 1 shows how to use a class to represent a complex data structure.
The class allows me to define a data structure with type annotations so that I can verify that Iâm using the data types correctly.
It does help to understand a bit about OOP.
For instance, classes define the attributes of an object, and classes can inherit attributes from parent classes, but this essentially describes the limits of how and why I use OOP in Python.
If you donât entirely follow that right now, donât worry.
Youâll understand it once you see it.
Instead of object-oriented code, I demonstrate programs composed almost entirely of functions. These functions are also pure in that they will only act on the values given to them. That is, pure functions never rely on some hidden, mutable state like global variables, and they will always return the same values given the same arguments. Additionally, every function will have an associated test that I can run to verify it behaves predictably. Itâs my opinion that this leads to shorter programs that are more transparent and easier to test than solutions written using OOP. You may disagree and are of course welcome to write your solutions using whatever style of programming you prefer, so long as they pass the tests. The Python Functional Programming HOWTO documentation makes a good case for why Python is suited for functional programming (FP).
Finally, the programs in this book also avoid the use of exceptions, which I think is appropriate for short programs you write for personal use.
Managing exceptions so that they donât interrupt the flow of a program adds another level of complexity that I feel detracts from oneâs ability to understand a program.
Iâm generally unhappy with how to write functions in Python that return errors.
Many people would raise an exception and let a try/catch block handle the mistakes.
If I feel an exception is warranted, I will often choose to not catch it, instead letting the program crash.
In this respect, Iâm following an idea from Joe Armstrong, the creator of the Erlang language, who said, âThe Erlang way is to write the happy path, and not write twisty little passages full of error correcting code.â
If you choose to write programs and modules for public release, you will need to learn much more about exceptions and error handling, but thatâs beyond the scope of this book.
Structure
The book is divided into two main parts. The first part tackles 14 of the programming challenges found at the Rosalind.info website.1 The second part shows more complicated programs that demonstrate other patterns or concepts I feel are important in bioinformatics. Every chapter of the book describes a coding challenge for you to write and provides a test suite for you to determine when youâve written a working program.
Although the âZen of Pythonâ says âThere should be oneâand preferably only oneâobvious way to do it,â I believe you can learn quite a bit by attempting many different approaches to a problem. Perl was my gateway into bioinformatics, and the Perl communityâs spirit of âThereâs More Than One Way To Do Itâ (TMTOWTDI) still resonates with me. I generally follow a theme-and-variations approach to each chapter, showing many solutions to explore different aspects of Python syntax and data structures.
Test-Driven Development
More than the act of testing, the act of designing tests is one of the best bug preventers known. The thinking that must be done to create a useful test can discover and eliminate bugs before they are codedâindeed, test-design thinking can discover and eliminate bugs at every stage in the creation of software, from conception to specification, to design, coding, and the rest.
Boris Beizer, Software Testing Techniques (Thompson Computer Press)
Underlying all my experimentation will be test suites that Iâll constantly run to ensure the programs continue to work correctly. Whenever I have the opportunity, I try to teach test-driven development (TDD), an idea explained in a book by that title written by Kent Beck (Addison-Wesley, 2002). TDD advocates writing tests for code before writing the code. The typical cycle involves the following:
-
Add a test.
-
Run all tests and see if the new test fails.
-
Write the code.
-
Run tests.
-
Refactor code.
-
Repeat.
In the bookâs GitHub repository, youâll find the tests for each program youâll write. Iâll explain how to run and write tests, and I hope by the end of the material youâll believe in the common sense and basic decency of using TDD. I hope that thinking about tests first will start to change the way you understand and explore coding.
Using the Command Line and Installing Python
My experience in bioinformatics has always been centered around the Unix command line. Much of my day-to-day work has been on some flavor of Linux server, stitching together existing command-line programs using shell scripts, Perl, and Python. While I might write and debug a program or a pipeline on my laptop, I will often deploy my tools to a high-performance compute (HPC) cluster where a scheduler will run my programs asynchronously, often in the middle of the night or over a weekend and without any supervision or intervention by me. Additionally, all my work building databases and websites and administering servers is done entirely from the command line, so I feel strongly that you need to master this environment to be successful in bioinformatics.
I used a Macintosh to write and test all the material for this book, and macOS has the Terminal app you can use for a command line.
I have also tested all the programs using various Linux distributions, and the GitHub repository includes instructions on how to use a Linux virtual machine with Docker.
Additionally, I tested all the programs on Windows 10 using the Ubuntu distribution Windows Subsystem for Linux (WSL) version 1.
I highly recommend WSL for Windows users to have a true Unix command line, but Windows shells like cmd.exe, PowerShell, and Git Bash can sometimes work sufficiently well for some programs.
I would encourage you to explore integrated development environments (IDEs) like VS Code, PyCharm, or Spyder to help you write, run, and test your programs.
These tools integrate text editors, help documentation, and terminals.
Although I wrote all the programs, tests, and even this book using the vim editor in a terminal, most people would probably prefer to use at least a more modern text editor like Sublime, TextMate, or Notepad++.
I wrote and tested all the examples using Python versions 3.8.6 and 3.9.1.
Some examples use Python syntax that was not present in version 3.6, so I would recommend you not use that version.
Python version 2.x is no longer supported and should not be used.
I tend to get the latest version of Python 3 from the Python download page, but Iâve also had success using the Anaconda Python distribution.
You may have a package manager like apt on Ubuntu or brew on Mac that can install a recent version, or you may choose to build from source.
Whatever your platform and installation method, I would recommend you try to use the most recent version as the language continues to change, mostly for the better.
Note that Iâve chosen to present the programs as command-line programs and not as Jupyter Notebooks for several reasons.
I like Notebooks for data exploration, but the source code for Notebooks is stored in JavaScript Object Notation (JSON) and not as line-oriented text.
This makes it very difficult to use tools like diff to find the differences between two Notebooks.
Also, Notebooks cannot be parameterized, meaning I cannot pass in arguments from outside the program to change the behavior but instead have to change the source code itself.
This makes the programs inflexible and automated testing impossible.
While I encourage you to explore Notebooks, especially as an interactive way to run Python, I will focus on how to write command-line programs.
Getting the Code and Tests
All the code and tests are available from the bookâs GitHub repository. You can use the program Git (which you may need to install) to copy the code to your computer with the following command. This will create a new directory called biofx_python on your computer with the contents of the repository:
$ git clone https://github.com/kyclark/biofx_python
If you enjoy using an IDE, it may be possible to clone the repository through that interface, as shown in Figure P-1. Many IDEs can help you manage projects and write code, but they all work differently. To keep things simple, I will show how to use the command line to accomplish most tasks.
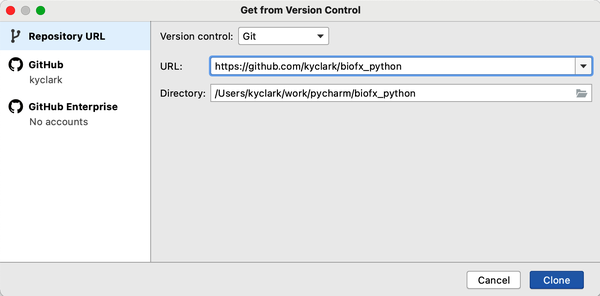
Figure P-1. The PyCharm tool can directly clone the GitHub repository for you
Some tools, like PyCharm, may automatically try to create a virtual environment inside the project directory. This is a way to insulate the version of Python and modules from other projects on your computer. Whether or not you use virtual environments is a personal preference. It is not a requirement to use them.
You may prefer to make a copy of the code in your own account so that you can track your changes and share your solutions with others. This is called forking because youâre breaking off from my code and adding your programs to the repository.
To fork my GitHub repository, do the following:
-
Create an account on GitHub.com.
-
Click the Fork button in the upper-right corner (see Figure P-2) to make a copy of the repository in your account.
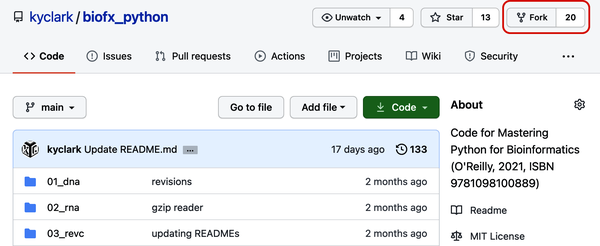
Figure P-2. The Fork button on my GitHub repository will make a copy of the code in your account
Now that you have a copy of all my code in your repository, you can use Git to copy that code to your computer.
Be sure to replace YOUR_GITHUB_ID with your actual GitHub ID:
$ git clone https://github.com/YOUR_GITHUB_ID/biofx_python
I may update the repo after you make your copy. If you would like to be able to get those updates, you will need to configure Git to set my repository as an upstream source. To do so, after you have cloned your repository to your computer, go into your biofx_python directory:
$ cd biofx_python
Then execute this command:
$ git remote add upstream https://github.com/kyclark/biofx_python.git
Whenever you would like to update your repository from mine, you can execute this command:
$ git pull upstream main
Installing Modules
You will need to install several Python modules and tools. Iâve included a requirements.txt file in the top level of the repository. This file lists all the modules needed to run the programs in the book. Some IDEs may detect this file and offer to install these for you, or you can use the following command:
$ python3 -m pip install -r requirements.txt
Or use the pip3 tool:
$ pip3 install -r requirements.txt
Sometimes pylint may complain about some of the variable names in the programs, and mypy will raise some issues when you import modules that do not have type annotations.
To silence these errors, you can create initialization files in your home directory that these programs will use to customize their behavior.
In the root of the source repository, there are files called pylintrc and mypy.ini that you should copy to your home directory like so:
$ cp pylintrc ~/.pylintrc $ cp mypy.ini ~/.mypy.ini
Alternatively, you can generate a new pylintrc with the following command:
$ cd ~ $ pylint --generate-rcfile > .pylintrc
Feel free to customize these files to suit your tastes.
Installing the new.py Program
I wrote a Python program called new.py that creates Python programs.
So meta, I know.
I wrote this for myself and then gave it to my students because I think itâs quite difficult to start writing a program from an empty screen.
The new.py program will create a new, well-structured Python program that uses the argparse module to interpret command-line arguments.
It should have been installed in the preceding section with the module dependencies.
If not, you can use the pip module to install it, like so:
$ python3 -m pip install new-py
You should now be able to execute new.py and see something like this:
$ new.py
usage: new.py [-h] [-n NAME] [-e EMAIL] [-p PURPOSE] [-t] [-f] [--version]
program
new.py: error: the following arguments are required: program
Each exercise will suggest that you use new.py to start writing your new programs.
For instance, in Chapter 1 you will create a program called dna.py in the 01_dna directory, like so:
$ cd 01_dna/ $ new.py dna.py Done, see new script "dna.py".
If you then execute ./dna.py --help, you will see that it generates help documentation on how to use the program.
You should open the dna.py program in your editor, modify the arguments, and add your code to satisfy the requirements of the program and the tests.
Note that itâs never a requirement that you use new.py.
I only offer this as an aid to getting started.
This is how I start every one of my own programs, but, while I find it useful, you may prefer to go a different route.
As long as your programs pass the test suites, you are welcome to write them however you please.
Why Did I Write This Book?
Richard Hamming spent decades as a mathematician and researcher at Bell Labs. He was known for seeking out people he didnât know and asking them about their research. Then he would ask them what they thought were the biggest, most pressing unanswered questions in their field. If their answers for both of these werenât the same, heâd ask, âSo why arenât you working on that?â
I feel that one of the most pressing problems in bioinformatics is that much of the software is poorly written and lacks proper documentation and testing, if it has any at all. I want to show you that itâs less difficult to use types and tests and linters and formatters because it will prove easier over time to add new features and release more and better software. You will have the confidence to know for certain when your program is correct, for at least some measure of correctness.
To that end, I will demonstrate best practices in software development. Though Iâm using Python as the medium, the principles apply to any language from C to R to JavaScript. The most important thing you can learn from this book is the craft of developing, testing, documenting, releasing, and supporting software, so that together we can all advance scientific research computing.
My career in bioinformatics was a product of wandering and happy accidents. I studied English literature and music in college, and then started playing with databases, HTML, and eventually learned programming on the job in the mid-1990s. By 2001, Iâd become a decent Perl hacker, and I managed to get a job as a web developer for Dr. Lincoln Stein, an author of several Perl modules and books, at Cold Spring Harbor Laboratory (CSHL). He and my boss, Dr. Doreen Ware, patiently spoon-fed me enough biology to understand the programs they wanted to be written. I spent 13 years working on a comparative plant genomics database called Gramene.org, learning a decent amount of science while continuing to explore programming languages and computer science.
Lincoln was passionate about sharing everything from data and code to education. He started the Programming for Biology course at CSHL, a two-week intensive crash course to teach Unix command-line, Perl programming, and bioinformatics skills. The course is still being taught, although using Python nowadays, and Iâve had several opportunities to act as a teaching assistant. Iâve always found it rewarding to help someone learn a skill they will use to further their research.
It was during my tenure at CSHL that I met Bonnie Hurwitz, who eventually left to pursue her PhD at the University of Arizona (UA). When she started her new lab at UA, I was her first hire. Bonnie and I worked together for several years, and teaching became one of my favorite parts of the job. As with Lincolnâs course, we introduced basic programming skills to scientists who wanted to branch out into more computational approaches.
Some of the materials I wrote for these classes became the foundation for my first book, Tiny Python Projects, where I try to teach the essential elements of Python language syntax as well as how to use tests to ensure that programs are correct and reproducibleâelements crucial to scientific programming. This book picks up from there and focuses on the elements of Python that will help you write programs for biology.
Conventions Used in This Book
The following typographical conventions are used in this book:
- Italic
-
Indicates new terms, URLs, email addresses, filenames, and file extensions, as well as codons and DNA bases.
Constant width-
Used for program listings, as well as within paragraphs to refer to program elements such as variable or function names, databases, data types, environment variables, statements, and keywords.
Constant width bold-
Shows commands or other text that should be typed literally by the user.
Constant width italic-
Shows text that should be replaced with user-supplied values or by values determined by context.
This element signifies a tip or suggestion.
This element signifies a general note.
This element indicates a warning or caution.
Using Code Examples
Supplemental material (code examples, exercises, etc.) is available for download at https://github.com/kyclark/biofx_python.
If you have a technical question or a problem using the code examples, please send email to bookquestions@oreilly.com.
This book is here to help you get your job done. In general, if example code is offered with this book, you may use it in your programs and documentation. You do not need to contact us for permission unless youâre reproducing a significant portion of the code. For example, writing a program that uses several chunks of code from this book does not require permission. Selling or distributing examples from OâReilly books does require permission. Answering a question by citing this book and quoting example code does not require permission. Incorporating a significant amount of example code from this book into your productâs documentation does require permission.
We appreciate, but generally do not require, attribution. An attribution usually includes the title, author, publisher, and ISBN. For example: âMastering Python for Bioinformatics by Ken Youens-Clark (OâReilly). Copyright 2021 Charles Kenneth Youens-Clark, 978-1-098-10088-9.â
If you feel your use of code examples falls outside fair use or the permission given above, feel free to contact us at permissions@oreilly.com.
OâReilly Online Learning
For more than 40 years, OâReilly Media has provided technology and business training, knowledge, and insight to help companies succeed.
Our unique network of experts and innovators share their knowledge and expertise through books, articles, and our online learning platform. OâReillyâs online learning platform gives you on-demand access to live training courses, in-depth learning paths, interactive coding environments, and a vast collection of text and video from OâReilly and 200+ other publishers. For more information, visit http://oreilly.com.
How to Contact Us
Please address comments and questions concerning this book to the publisher:
- OâReilly Media, Inc.
- 1005 Gravenstein Highway North
- Sebastopol, CA 95472
- 800-998-9938 (in the United States or Canada)
- 707-829-0515 (international or local)
- 707-829-0104 (fax)
We have a web page for this book, where we list errata, examples, and any additional information. You can access this page at https://oreil.ly/mastering-bioinformatics-python.
Email bookquestions@oreilly.com to comment or ask technical questions about this book.
For news and information about our books and courses, visit http://oreilly.com.
Find us on Facebook: http://facebook.com/oreilly
Follow us on Twitter: http://twitter.com/oreillymedia
Watch us on YouTube: http://www.youtube.com/oreillymedia
Acknowledgments
I want to thank the many people who have reviewed this book, including my editor, Corbin Collins; the entire production team but especially my production editor, Caitlin Ghegan; my technical reviewers, Al Scherer, Brad Fulton, Bill Lubanovic, Rangarajan Janani, and Joshua Orvis; and the many other people who provided much-appreciated feedback, including Mark Henderson, Marc Bañuls Tornero, and Dr. Scott Cain.
In my professional career, Iâve been extremely fortunate to have had many wonderful bosses, supervisors, and colleagues whoâve helped me grow and pushed me to be better. Eric Thorsen was the first person to see I had the potential to learn how to code, and he helped me learn various languages and databases as well as important lessons about sales and support. Steve Reppucci was my boss at boston.com, and he provided a much deeper understanding of Perl and Unix and how to be an honest and thoughtful team leader. Dr. Lincoln Stein at CSHL took a chance to hire someone who had no knowledge of biology to work in his lab, and he pushed me to create programs I didnât imagine I could. Dr. Doreen Ware patiently taught me biology and pushed me to assume leadership roles and publish. Dr. Bonnie Hurwitz supported me through many years of learning about high-performance computing, more programming languages, mentoring, teaching, and writing. In every position, I also had many colleagues who taught me as much about programming as about being human, and I thank everyone who has helped me along the way.
In my personal life, I would be nowhere without my family, who have loved and supported me. My parents have shown great support throughout my life, and I surely wouldnât be the person I am without them. Lori Kindler and I have been married 25 years, and I canât imagine a life without her. Together we generated three offspring who have been an incredible source of delight and challenge.
1 Named for Rosalind Franklin, who should have received a Nobel Prize for her contributions to discovering the structure of DNA.
Get Mastering Python for Bioinformatics now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

