Chapter 4. Data Ingestion: Extracting Data
As discussed in Chapter 3, the ELT pattern is the ideal design for data pipelines built for data analysis, data science, and data products. The first two steps in the ELT pattern, extract and load, are collectively referred to as data ingestion. This chapter discusses getting your development environment and infrastructure set up for both, and it goes through the specifics of extracting data from various source systems. Chapter 5 discusses loading the resulting datasets into a data warehouse.
Note
The extract and load code samples in this chapter are fully decoupled from each other. Coordinating the two steps to complete a data ingestion is a topic that’s discussed in Chapter 7.
As discussed in Chapter 2, there are numerous types of source systems to extract from, as well as numerous destinations to load into. In addition, data comes in many forms, all of which present different challenges for ingesting it.
This chapter and the next include code samples for exporting and ingesting data from and to common systems. The code is highly simplified and contains only minimal error handing. Each example is intended as an easy-to-understand starting point for data ingestions, but is fully functional and extendable to more scalable solutions.
Note
The code samples in this chapter write extracted data to CSV files to be loaded into the destination data warehouse. There are times when it makes more sense to store extracted data in another format, such as JSON, prior to loading. Where applicable, I note where you might want to consider making such an adjustment.
Chapter 5 also discusses some open source frameworks you can build off of, and commercial alternatives that give data engineers and analysts “low code” options for ingesting data.
Setting Up Your Python Environment
All code samples that follow are written in Python and SQL and use open source frameworks that are common in the data engineering field today. For simplicity, the number of sources and destinations is limited. However, where applicable, I provide notes on how to modify for similar systems.
To run the sample code, you’ll need a physical or virtual machine running Python 3.x. You’ll also need to install and import a few libraries.
If you don’t have Python installed on your machine, you can get the distribution and installer for your OS directly from them.
Note
The following commands are written for a Linux or Macintosh command line. On Windows, you may need to add the Python 3 executable to your PATH.
Before you install the libraries used in this chapter, it’s best to create a virtual environment to install them into. To do so, you can use a tool called virtualenv. virtualenv is helpful in managing Python libraries for different projects and applications. It allows you to install Python libraries within a scope specific to your project rather than globally. First, create a virtual environment named env.
$ python -m venv env
Now that your virtual environment is created, activate it with the following command:
$ source env/bin/activate
You can verify that your virtual environment is activated in two ways. First, you’ll notice that your command prompt is now prefixed by the environment name:
(env) $
You can also use the which python command to verify where Python is looking for libraries. You should see something like this, which shows the path of the virtual environment directory:
(env) $ which python env/bin/python
Now it’s safe to install the libraries you need for the code samples that follow.
Note
On some operating systems (OS), you must use python3 instead of python to run the Python 3.x executable. Older OS versions may default to Python 2.x. You can find out which version of Python your OS uses by typing python --version.
Throughout this chapter, you’ll use pip to install the libraries used in the code samples. pip is a tool that ships with most Python distributions.
The first library you’ll install using pip is configparser, which will be used to read configuration information you’ll add to a file later.
(env) $ pip install configparser
Next, create a file named pipeline.conf in the same directory as the Python scripts you’ll create in the following sections. Leave the file empty for now. The code samples in this chapter will call for adding to it. In Linux and Mac operating systems, you can create the empty file on the command line with the following command:
(env) $ touch pipeline.conf
Setting Up Cloud File Storage
For each example in this chapter, you’ll be using an Amazon Simple Storage Service (Amazon S3 or simply S3) bucket for file storage. S3 is hosted on AWS, and as the name implies, S3 is a simple way to store and access files. It’s also very cost effective. As of this writing, AWS offers 5 GB of free S3 storage for 12 months with a new AWS account and charges less than 3 cents USD per month per gigabyte for the standard S3 class of storage after that. Given the simplicity of the samples in this chapter, you’ll be able to store the necessary data in S3 for free if you are still in the first 12 months of creating an AWS account, or for less than a $1 a month after that.
To run the samples in this chapter, you’ll need an S3 bucket. Thankfully, creating an S3 bucket is simple, and the latest instructions can be found in the AWS documentation. Setting up the proper access control to the S3 bucket is dependent upon which data warehouse you are using. In general, it’s best to use AWS Identity and Access Management (IAM) roles for access management policies. Detailed instructions for setting up such access for both an Amazon Redshift and Snowflake data warehouse are in the sections that follow, but for now, follow the instruction to create a new bucket. Name it whatever you’d like; I suggest using the default settings, including keeping the bucket private.
Each extraction example extracts data from the given source system and stores the output in the S3 bucket. Each loading example in Chapter 5 loads that data from the S3 bucket into the destination. This is a common pattern in data pipelines. Every major public cloud provider has a service similar to S3. Equivalents on other public clouds are Azure Storage in Microsoft Azure and Google Cloud Storage (GCS) in GCP.
It’s also possible to modify each example to use local or on-premises storage. However, there is additional work required to load data into your data warehouse from storage outside of its specific cloud provider. Regardless, the patterns described in this chapter are valid no matter which cloud provider you use, or if you choose to host your data infrastructure on-premises.
Before I move on to each example, there’s one more Python library that you’ll need to install so that your scripts for extracting and loading can interact with your S3 bucket. Boto3 is the AWS SDK for Python. Make sure the virtual environment you set up in the previous section is active and use pip to install it:
(env) $ pip install boto3
In the examples that follow, you’ll be asked to import boto3 into your Python scripts like this:
importboto3
Because you’ll be using the boto3 Python library to interact with your S3 bucket, you’ll also need to create an IAM user, generate access keys for that user, and store the keys in a configuration file that your Python scripts can read from. This is all necessary so that your scripts have permissions to read and write files in your S3 bucket.
First, create the IAM user:
-
Under the Services menu in the AWS console (or top nav bar), navigate to IAM.
-
In the navigation pane, click Users and then click “Add user.” Type the username for the new user. In this example, name the user data_pipeline_readwrite.
-
Click the type of access for this IAM user. Click “programmatic access” since this user won’t need to log into the AWS Console, but rather access AWS resources programmatically via Python scripts.
-
Click Next: Permissions.
-
On the “Set permissions” page, click the “Attach existing policies to user directly” option. Add the AmazonS3FullAccess policy.
-
Click Next: Tags. It’s a best practice in AWS to add tags to various objects and services so you can find them later. This is optional, however.
-
Click Next: Review to verify your settings. If everything looks good, click “Create user.”
-
You’ll want to save the access key ID and secret access key for the new IAM user. To do so, click Download.csv and then save the file to a safe location so you can use it in just a moment.
Finally, add a section to the pipeline.conf file called [aws_boto_credentials] to store the credentials for the IAM user and the S3 bucket information. You can find your AWS account ID by clicking your account name at the top right of any page when logged into the AWS site. Use the name of the S3 bucket you created earlier for the bucket_name value. The new section in pipline.conf will look like this:
[aws_boto_credentials] access_key = ijfiojr54rg8er8erg8erg8 secret_key = 5r4f84er4ghrg484eg84re84ger84 bucket_name = pipeline-bucket account_id = 4515465518
Extracting Data from a MySQL Database
Extracting data from a MySQL database can be done in two ways:
-
Full or incremental extraction using SQL
-
Binary Log (binlog) replication
Full or incremental extraction using SQL is far simpler to implement, but also less scalable for large datasets with frequent changes. There are also trade-offs between full and incremental extractions that I discuss in the following section.
Binary Log replication, though more complex to implement, is better suited to cases where the data volume of changes in source tables is high, or there is a need for more frequent data ingestions from the MySQL source.
Note
Binlog replication is also a path to creating a streaming data ingestion. See the “Batch Versus Stream Ingestion” section of this chapter for more on the distinction between the two approaches as well as implementation patterns.
This section is relevant to those readers who have a MySQL data source they need to extract data from. However, if you’d like to set up a simple database so you can try the code samples, you have two options. First, you can install MySQL on your local machine or virtual machine for free. You can find an installer for your OS on the MySQL downloads page.
Alternatively, you can create a fully managed Amazon RDS for MySQL instance in AWS. I find this method more straightforward, and it’s nice not to create unnecessary clutter on my local machine!
Warning
When you follow the linked instructions to set up an MySQL RDS database instance, you’ll be prompted to set your database as publicly accessible. That’s just fine for learning and working with sample data. In fact, it makes it much easier to connect from whatever machine you’re running the samples in this section. However, for more robust security in a production setting, I suggest following the Amazon RDS security best practices.
Note that just like the S3 pricing noted earlier, if you are no longer eligible for the free tier of AWS, there is a cost associated with doing so. Otherwise, it’s free to set up and run! Just remember to delete your RDS instance when you’re done so you don’t forget and incur charges when your free tier expires.
The code samples in this section are quite simple and refer to a table named Orders in a MySQL database. Once you have a MySQL instance to work with, you can create the table and insert some sample rows by running the following SQL commands:
CREATETABLEOrders(OrderIdint,OrderStatusvarchar(30),LastUpdatedtimestamp);INSERTINTOOrdersVALUES(1,'Backordered','2020-06-01 12:00:00');INSERTINTOOrdersVALUES(1,'Shipped','2020-06-09 12:00:25');INSERTINTOOrdersVALUES(2,'Shipped','2020-07-11 3:05:00');INSERTINTOOrdersVALUES(1,'Shipped','2020-06-09 11:50:00');INSERTINTOOrdersVALUES(3,'Shipped','2020-07-12 12:00:00');
Full or Incremental MySQL Table Extraction
When you need to ingest either all or a subset of columns from a MySQL table into a data warehouse or data lake, you can do so using either full extraction or incremental extraction.
In a full extraction, every record in the table is extracted on each run of the extraction job. This is the least complex approach, but for high-volume tables it can take a long time to run. For example, if you want to run a full extraction on a table called Orders, the SQL executed on the source MySQL database will look like this:
SELECT*FROMOrders;
In an incremental extraction, only records from the source table that have changed or been added since the last run of the job are extracted. The timestamp of the last extraction can either be stored in an extraction job log table in the data warehouse or retrieved by querying the maximum timestamp in a LastUpdated column in the destination table in the warehouse. Using the fictional Orders table as an example, the SQL query executed on the source MySQL database will look like this:
SELECT*FROMOrdersWHERELastUpdated>{{last_extraction_run}};
Note
For tables containing immutable data (meaning records can be inserted, but not updated), you can make use of the timestamp for when the record was created instead of a LastUpdated column.
The {{ last_extraction_run }} variable is a timestamp representing the most recent run of the extraction job. Most commonly it’s queried from the destination table in the data warehouse. In that case, the following SQL would be executed in the data warehouse, with the resulting value used for {{ last_extraction_run }}:
SELECTMAX(LastUpdated)FROMwarehouse.Orders;
Though incremental extraction is ideal for optimal performance, there are some downsides and reasons why it may not be possible for a given table.
First, with this method deleted, rows are not captured. If a row is deleted from the source MySQL table, you won’t know, and it will remain in the destination table as if nothing changed.
Second, the source table must have a reliable timestamp for when it was last updated (the LastUpdated column in the previous example). It’s not uncommon for source system tables to be missing such a column or have one that is not updated reliably. There’s nothing stopping developers from updating records in the source table and forgetting to update the LastUpdated timestamp.
However, incremental extraction does make it easier to capture updated rows. In the upcoming code samples, if a particular row in the Orders table is updated, both the full and incremental extractions will bring back the latest version of the row. In the full extract, that’s true for all rows in the table as the extraction retrieves a full copy of the table. In the incremental extraction, only rows that have changed are retrieved.
When it comes time for the load step, full extracts are usually loaded by first truncating the destination table and loading in the newly extracted data. In that case, you’re left with only the latest version of the row in the data warehouse.
When loading data from an incremental extraction, the resulting data is appended to the data in the destination table. In that case, you have both the original record as well as the updated version. Having both can be valuable when it comes time to transform and analyze data, as I discuss in Chapter 6.
For example, Table 4-1 shows the original record for OrderId 1 in the MySQL database. When the order was placed by the customer, it was on back order. Table 4-2 shows the updated record in the MySQL database. As you can see, the order was updated because it shipped on 2020-06-09.
| OrderId | OrderStatus | LastUpdated |
|---|---|---|
1 |
Backordered |
2020-06-01 12:00:00 |
| OrderId | OrderStatus | LastUpdated |
|---|---|---|
1 |
Shipped |
2020-06-09 12:00:25 |
When a full extraction is run, the destination table in the data warehouse is first truncated and then loaded with the output of the extraction. The result for OrderId 1 is the single record shown in Table 4-2. In an incremental extraction, however, the output of the extract is simply appended to the destination table in the data warehouse. The result is both the original and updated records for OrderId 1 being in the data warehouse, as illustrated in Table 4-3.
| OrderId | OrderStatus | LastUpdated |
|---|---|---|
1 |
Backordered |
2020-06-01 12:00:00 |
1 |
Shipped |
2020-06-09 12:00:25 |
You can learn more about loading full and incremental extractions in sections of Chapter 5 including “Loading Data into a Redshift Warehouse”.
Warning
Never assume a LastUpdated column in a source system is reliably updated. Check with the owner of the source system and confirm before relying on it for an incremental extraction.
Both full and incremental extractions from a MySQL database can be implemented using SQL queries executed on the database but triggered by Python scripts. In addition to the Python libraries installed in previous sections, you’ll need to install the PyMySQL library using pip:
(env) $ pip install pymysql
You’ll also need to add a new section to the pipeline.conf file to store the connection information for the MySQL database:
[mysql_config] hostname = my_host.com port = 3306 username = my_user_name password = my_password database = db_name
Now create a new Python script named extract_mysql_full.py. You’ll need to import several libraries, such as pymysql, which connects to the MySQL database, and the csv library so that you can structure and write out the extracted data in a flat file that’s easy to import into a data warehouse in the load step of ingestion. Also, import boto3 so that you can upload the resulting CSV file to your S3 bucket for later loading into the data warehouse:
importpymysqlimportcsvimportboto3importconfigparser
Now you can initialize a connection to the MySQL database:
parser=configparser.ConfigParser()parser.read("pipeline.conf")hostname=parser.get("mysql_config","hostname")port=parser.get("mysql_config","port")username=parser.get("mysql_config","username")dbname=parser.get("mysql_config","database")password=parser.get("mysql_config","password")conn=pymysql.connect(host=hostname,user=username,password=password,db=dbname,port=int(port))ifconnisNone:("Error connecting to the MySQL database")else:("MySQL connection established!")
Run a full extraction of the Orders table from the earlier example. The following code will extract the entire contents of the table and write it to a pipe-delimited CSV file. To perform the extraction, it uses a cursor object from the pymysql library to execute the SELECT query:
m_query="SELECT * FROM Orders;"local_filename="order_extract.csv"m_cursor=conn.cursor()m_cursor.execute(m_query)results=m_cursor.fetchall()withopen(local_filename,'w')asfp:csv_w=csv.writer(fp,delimiter='|')csv_w.writerows(results)fp.close()m_cursor.close()conn.close()
Now that the CSV file is written locally, it needs to be uploaded to the S3 bucket for later loading into the data warehouse or other destination. Recall from “Setting Up Cloud File Storage” that you set up an IAM user for the Boto3 library to use for authentication to the S3 bucket. You also stored the credentials in the aws_boto_credentials section of the pipeline.conf file. Here is the code to upload the CSV file to your S3 bucket:
# load the aws_boto_credentials valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")access_key=parser.get("aws_boto_credentials","access_key")secret_key=parser.get("aws_boto_credentials","secret_key")bucket_name=parser.get("aws_boto_credentials","bucket_name")s3=boto3.client('s3',aws_access_key_id=access_key,aws_secret_access_key=secret_key)s3_file=local_filenames3.upload_file(local_filename,bucket_name,s3_file)
You can execute the script as follows:
(env) $ python extract_mysql_full.py
When the script is executed, the entire contents of the Orders table is now contained in a CSV file sitting in the S3 bucket waiting to be loaded into the data warehouse or other data store. See Chapter 5 for more on loading into the data store of your choice.
If you want to extract data incrementally, you’ll need to make a few changes to the script. I suggest creating a copy of extract_mysql_full.py named extract_mysql_incremental.py as a starting point.
First, find the timestamp of the last record that was extracted from the source Orders table. To do that, query the MAX(LastUpdated) value from the Orders table in the data warehouse. In this example, I’ll use a Redshift data warehouse (see “Configuring an Amazon Redshift Warehouse as a Destination”), but you can use the same logic with the warehouse of your choice.
To interact with your Redshift cluster, install the psycopg2 library, if you haven’t already.
(env) $ pip install psycopg2
Here is the code to connect to and query the Redshift cluster to get the MAX(LastUpdated) value from the Orders table:
importpsycopg2# get db Redshift connection infoparser=configparser.ConfigParser()parser.read("pipeline.conf")dbname=parser.get("aws_creds","database")user=parser.get("aws_creds","username")password=parser.get("aws_creds","password")host=parser.get("aws_creds","host")port=parser.get("aws_creds","port")# connect to the redshift clusterrs_conn=psycopg2.connect("dbname="+dbname+" user="+user+" password="+password+" host="+host+" port="+port)rs_sql="""SELECT COALESCE(MAX(LastUpdated),'1900-01-01')FROM Orders;"""rs_cursor=rs_conn.cursor()rs_cursor.execute(rs_sql)result=rs_cursor.fetchone()# there's only one row and column returnedlast_updated_warehouse=result[0]rs_cursor.close()rs_conn.commit()
Using the value stored in last_updated_warehouse, modify the extraction query run on the MySQL database to pull only those records from the Orders table that have been updated since the prior run of the extraction job. The new query contains a placeholder, represented by %s for the last_updated_warehouse value. The value is then passed into the cursor’s .execute() function as a tuple (a data type used to store collections of data). This is the proper and secure way to add parameters to a SQL query to avoid possible SQL injection. Here is the updated code block for running the SQL query on the MySQL database:
m_query="""SELECT *FROM OrdersWHERE LastUpdated >%s;"""local_filename="order_extract.csv"m_cursor=conn.cursor()m_cursor.execute(m_query,(last_updated_warehouse,))
The entire extract_mysql_incremental.py script for the incremental extraction (using a Redshift cluster for the last_updated value) looks like this:
importpymysqlimportcsvimportboto3importconfigparserimportpsycopg2# get db Redshift connection infoparser=configparser.ConfigParser()parser.read("pipeline.conf")dbname=parser.get("aws_creds","database")user=parser.get("aws_creds","username")password=parser.get("aws_creds","password")host=parser.get("aws_creds","host")port=parser.get("aws_creds","port")# connect to the redshift clusterrs_conn=psycopg2.connect("dbname="+dbname+" user="+user+" password="+password+" host="+host+" port="+port)rs_sql="""SELECT COALESCE(MAX(LastUpdated),'1900-01-01')FROM Orders;"""rs_cursor=rs_conn.cursor()rs_cursor.execute(rs_sql)result=rs_cursor.fetchone()# there's only one row and column returnedlast_updated_warehouse=result[0]rs_cursor.close()rs_conn.commit()# get the MySQL connection info and connectparser=configparser.ConfigParser()parser.read("pipeline.conf")hostname=parser.get("mysql_config","hostname")port=parser.get("mysql_config","port")username=parser.get("mysql_config","username")dbname=parser.get("mysql_config","database")password=parser.get("mysql_config","password")conn=pymysql.connect(host=hostname,user=username,password=password,db=dbname,port=int(port))ifconnisNone:("Error connecting to the MySQL database")else:("MySQL connection established!")m_query="""SELECT *FROM OrdersWHERE LastUpdated >%s;"""local_filename="order_extract.csv"m_cursor=conn.cursor()m_cursor.execute(m_query,(last_updated_warehouse,))results=m_cursor.fetchall()withopen(local_filename,'w')asfp:csv_w=csv.writer(fp,delimiter='|')csv_w.writerows(results)fp.close()m_cursor.close()conn.close()# load the aws_boto_credentials valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")access_key=parser.get("aws_boto_credentials","access_key")secret_key=parser.get("aws_boto_credentials","secret_key")bucket_name=parser.get("aws_boto_credentials","bucket_name")s3=boto3.client('s3',aws_access_key_id=access_key,aws_secret_access_key=secret_key)s3_file=local_filenames3.upload_file(local_filename,bucket_name,s3_file)
Warning
Beware of large extraction jobs—whether full or incremental—putting strain on the source MySQL database, and even blocking production queries from executing. Consult with the owner of the database and consider setting up a replica to extract from, rather than extracting from the primary source database.
Binary Log Replication of MySQL Data
Though more complex to implement, ingesting data from a MySQL database using the contents of the MySQL binlog to replicate changes is efficient in cases of high-volume ingestion needs.
Note
Binlog replication is a form of change data capture (CDC). Many source data stores have some form of CDC that you can use.
The MySQL binlog is a log that keeps a record of every operation performed in the database. For example, depending on how it’s configured, it will log the specifics of every table creation or modification, as well as every INSERT, UPDATE, and DELETE operation. Though originally intended to replicate data to other MySQL instances, it’s not hard to see why the contents of the binlog are so appealing to data engineers who want to ingest data into a data warehouse.
Because your data warehouse is likely not a MySQL database, it’s not possible to simply use the built-in MySQL replication features. To make use of the binlog for data ingestion to a non-MySQL source, there are a number of steps to take:
-
Enable and configure the binlog on the MySQL server.
-
Run an initial full table extraction and load.
-
Extract from the binlog on a continuous basis.
-
Translate and load binlog extracts into the data warehouse.
Note
Step 3 is not discussed in detail, but to use the binlog for ingestion, you must first populate the tables in the data warehouse with the current state of the MySQL database and then use the binlog to ingest subsequent changes. Doing so often involves putting a LOCK on the tables you want to extract, running a mysqldump of those tables, and then loading the result of the mysqldump into the warehouse before turning on the binlog ingestion.
Though it’s best to refer to the latest MySQL binlog documentation for instructions in enabling and configuring binary logging, I will walk through the key configuration values.
There are two key settings to ensure on the MySQL database in regard to binlog configuration.
First, ensure that binary logging is enabled. Typically it is enabled by default, but you can check by running the following SQL query on the database (exact syntax may vary by MySQL distribution):
SELECTvariable_valueasbin_log_statusFROMperformance_schema.global_variablesWHEREvariable_name='log_bin';
If the binary logging is enabled, you’ll see the following. If the status returned is OFF, then you’ll need to consult the MySQL documentation for the relevant version to enable it.
+ — — — — — — — — — — — — — — — — — — -+ | bin_log_status :: | + — — — — — — — — — — — — — — — — — — -+ | ON | + — — — — — — — — — — — — — — — — — — -+ 1 row in set (0.00 sec)
Next, ensure that the binary logging format is set appropriately. There are three formats supported in the recent version of MySQL:
-
STATEMENT -
ROW -
MIXED
The STATEMENT format logs every SQL statement that inserts or modifies a row in the binlog. If you wanted to replicate data from one MySQL database to another, this format is useful. To replicate the data, you could just run all statements to reproduce the state of the database. However, because the extracted data is likely bound for a data warehouse running on a different platform, the SQL statements produced in the MySQL database may not be compatible with your data warehouse.
With the ROW format, every change to a row in a table is represented on a line of the binlog not as a SQL statement but rather the data in the row itself. This is the preferred format to use.
The MIXED format logs both STATEMENT- and ROW-formatted records in the binlog. Though it’s possible to sift out just the ROW data later, unless the binlog is being used for another purpose, it’s not necessary to enable MIXED, given the additional disk space that it takes up.
You can verify the current binlog format by running the following SQL query:
SELECTvariable_valueasbin_log_formatFROMperformance_schema.global_variablesWHEREvariable_name='binlog_format';
The statement will return the format that’s currently active:
+ — — — — — — — — — — — — — — — — — — — -+ | bin_log_format :: | + — — — — — — — — — — — — — — — — — — — -+ | ROW | + — — — — — — — — — — — — — — — — — — — -+ 1 row in set (0.00 sec)
The binlog format as well as other configuration settings are typically set in the my.cnf file specific to the MySQL database instance. If you open the file, you’ll see a row like the following included:
[mysqld] binlog_format=row ........
Again, it’s best to consult with the owner of the MySQL database or the latest MySQL documentation before modifying any configurations.
Now that binary logging is enabled in a ROW format, you can build a process to extract the relevant information from it and store it in a file to be loaded into your data warehouse.
There are three different types of ROW-formatted events that you’ll want to pull from the binlog. For the sake of this ingestion example, you can ignore other events you find in the log, but in more advanced replication strategies, extracting events that modify the structure of a table is also of value. The events that you’ll work with are as follows:
-
WRITE_ROWS_EVENT -
UPDATE_ROWS_EVENT -
DELETE_ROWS_EVENT
Next, it’s time to get the events from the binlog. Thankfully, there are some open source Python libraries available to get you started. One of the most popular is the python-mysql-replication project, which can be found on GitHub. To get started, install it using pip:
(env) $ pip install mysql-replication
To get an idea of what the output from the binlog looks like, you can connect to the database and read from the binlog. In this example, I’ll use the MySQL connection information added to the pipeline.conf file for the full and incremental ingestion example earlier in this section.
Note
The following example reads from the MySQL server’s default binlog file. The default binlog filename and path are set in the log_bin variable, which is stored in the my.cnf file for the MySQL database. In some cases, binlogs are rotated over time (perhaps daily or hourly). If so, you will need to determine the file path based on the method of log rotation and file naming scheme chosen by the MySQL administrator and pass it as a value to the log_file parameter when creating the BinLogStreamReader instance. See the documentation for the BinLogStreamReader class for more.
frompymysqlreplicationimportBinLogStreamReaderfrompymysqlreplicationimportrow_eventimportconfigparserimportpymysqlreplication# get the MySQL connection infoparser=configparser.ConfigParser()parser.read("pipeline.conf")hostname=parser.get("mysql_config","hostname")port=parser.get("mysql_config","port")username=parser.get("mysql_config","username")password=parser.get("mysql_config","password")mysql_settings={"host":hostname,"port":int(port),"user":username,"passwd":password}b_stream=BinLogStreamReader(connection_settings=mysql_settings,server_id=100,only_events=[row_event.DeleteRowsEvent,row_event.WriteRowsEvent,row_event.UpdateRowsEvent])foreventinb_stream:event.dump()b_stream.close()
There are a few things to note about the BinLogStreamReader object that’s instantiated in the code sample. First, it connects to the MySQL database specified in the pipeline.conf file and reads from a specific binlog file. Next, the combination of the resume_stream=True setting and the log_pos value tells it to start reading the binlog at a specified point. In this case, that’s position 1400. Finally, I tell BinLogStreamReader to only read the DeleteRowsEvent, WriteRowsEvent, and UpdateRowsEvent, events using the only_events parameter.
Next, the script iterates through all of the events and prints them in a human-readable format. For your database with the Orders table in it, you’ll see something like this as output:
=== WriteRowsEvent === Date: 2020-06-01 12:00:00 Log position: 1400 Event size: 30 Read bytes: 20 Table: orders Affected columns: 3 Changed rows: 1 Values: -- * OrderId : 1 * OrderStatus : Backordered * LastUpdated : 2020-06-01 12:00:00 === UpdateRowsEvent === Date: 2020-06-09 12:00:25 Log position: 1401 Event size: 56 Read bytes: 15 Table: orders Affected columns: 3 Changed rows: 1 Affected columns: 3 Values: -- * OrderId : 1 => 1 * OrderStatus : Backordered => Shipped * LastUpdated : 2020-06-01 12:00:00 => 2020-06-09 12:00:25
As you can see, there are two events that represent the INSERT and UPDATE of OrderId 1, which was shown in Table 4-3. In this fictional example, the two sequential binlog events are days apart, but in reality there would be numerous events between them, representing all changes made in the database.
Note
The value of log_pos, which tells BinLogStreamReader where to start, is a value that you’ll need to store somewhere in a table of your own to keep track of where to pick up when the next extract runs. I find it best to store the value in a log table in the data warehouse from which it can be read when the extraction starts and to which it can be written, with the position value of the final event when it finishes.
Though the code sample shows what the events look like in a human-readable format, to make the output easy to load into the data warehouse, it’s necessary to do a couple more things:
-
Parse and write the data in a different format. To simplify loading, the next code sample will write each event to a row in a CSV file.
-
Write one file per table that you want to extract and load. Though the example binlog only contains events related to the
Orderstable, it’s highly likely that in a real binlog, events related to other tables are included as well.
To address the first change, instead of using the .dump() function I will instead parse out the event attributes and write them to a CSV file. For the second, instead of writing a file for each table, for simplicity I will only write events related to the Orders table to a file called orders_extract.csv. In a fully implemented extraction, modify this code sample to group events by table and write multiple files, one for each table you want to ingest changes for. The last step in the final code sample uploads the CSV file to the S3 bucket so it can be loaded into the data warehouse, as described in detail in Chapter 5:
frompymysqlreplicationimportBinLogStreamReaderfrompymysqlreplicationimportrow_eventimportconfigparserimportpymysqlreplicationimportcsvimportboto3# get the MySQL connection infoparser=configparser.ConfigParser()parser.read("pipeline.conf")hostname=parser.get("mysql_config","hostname")port=parser.get("mysql_config","port")username=parser.get("mysql_config","username")password=parser.get("mysql_config","password")mysql_settings={"host":hostname,"port":int(port),"user":username,"passwd":password}b_stream=BinLogStreamReader(connection_settings=mysql_settings,server_id=100,only_events=[row_event.DeleteRowsEvent,row_event.WriteRowsEvent,row_event.UpdateRowsEvent])order_events=[]forbinlogeventinb_stream:forrowinbinlogevent.rows:ifbinlogevent.table=='orders':event={}ifisinstance(binlogevent,row_event.DeleteRowsEvent):event["action"]="delete"event.update(row["values"].items())elifisinstance(binlogevent,row_event.UpdateRowsEvent):event["action"]="update"event.update(row["after_values"].items())elifisinstance(binlogevent,row_event.WriteRowsEvent):event["action"]="insert"event.update(row["values"].items())order_events.append(event)b_stream.close()keys=order_events[0].keys()local_filename='orders_extract.csv'withopen(local_filename,'w',newline='')asoutput_file:dict_writer=csv.DictWriter(output_file,keys,delimiter='|')dict_writer.writerows(order_events)# load the aws_boto_credentials valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")access_key=parser.get("aws_boto_credentials","access_key")secret_key=parser.get("aws_boto_credentials","secret_key")bucket_name=parser.get("aws_boto_credentials","bucket_name")s3=boto3.client('s3',aws_access_key_id=access_key,aws_secret_access_key=secret_key)s3_file=local_filenames3.upload_file(local_filename,bucket_name,s3_file)
After execution, orders_extract.csv will look like this:
insert|1|Backordered|2020-06-01 12:00:00 update|1|Shipped|2020-06-09 12:00:25
As I discuss in Chapter 5, the format of the resulting CSV file is optimized for fast loading. Making sense of the data that’s been extracted is a job for the transform step in a pipeline, reviewed in detail in Chapter 6.
Extracting Data from a PostgreSQL Database
Just like MySQL, ingesting data from a PostgreSQL (commonly known as Postgres) database can be done in one of two ways: either with full or incremental extractions using SQL or by leveraging features of the database meant to support replication to other nodes. In the case of Postgres, there are a few ways to do this, but this chapter will focus on one method: turning the Postgres write-ahead log (WAL) into a data stream.
Like the previous section, this one is intended for those who need to ingest data from an existing Postgres database. However, if you’d like to just try the code samples, you can set up Postgres either by installing on your local machine, or in AWS by using an a RDS instance, which I recommend. See the previous section for notes on pricing and security-related best practices for RDS MySQL, as they apply to RDS Postgres as well.
The code samples in this section are quite simple and refer to a table named Orders in a Postgres database. Once you have a Postgres instance to work with, you can create the table and insert some sample rows by running the following SQL commands:
CREATETABLEOrders(OrderIdint,OrderStatusvarchar(30),LastUpdatedtimestamp);INSERTINTOOrdersVALUES(1,'Backordered','2020-06-01 12:00:00');INSERTINTOOrdersVALUES(1,'Shipped','2020-06-09 12:00:25');INSERTINTOOrdersVALUES(2,'Shipped','2020-07-11 3:05:00');INSERTINTOOrdersVALUES(1,'Shipped','2020-06-09 11:50:00');INSERTINTOOrdersVALUES(3,'Shipped','2020-07-12 12:00:00');
Full or Incremental Postgres Table Extraction
This method is similar to full and incremental and full extractions demonstrated in “Extracting Data from a MySQL Database”. It’s so similar that I won’t go into detail here beyond one difference in the code. Like the example in that section, this one will extract data from a table called Orders in a source database, write it to a CSV file, and then upload it to an S3 bucket.
The only difference in this section is the Python library I’ll use to extract the data. Instead of PyMySQL, I’ll be using pyscopg2 to connect to a Postgres database. If you have not already installed it, you can do so using pip:
(env) $ pip install pyscopg2
You’ll also need to add a new section to the pipeline.conf file with the connection information for the Postgres database:
[postgres_config] host = myhost.com port = 5432 username = my_username password = my_password database = db_name
The code to run the full extraction of the Orders table is nearly identical to the sample from the MySQL section, but as you can see, it uses pyscopg2 to connect to the source database and to run the query. Here it is in its entirety:
importpsycopg2importcsvimportboto3importconfigparserparser=configparser.ConfigParser()parser.read("pipeline.conf")dbname=parser.get("postgres_config","database")user=parser.get("postgres_config","username")password=parser.get("postgres_config","password")host=parser.get("postgres_config","host")port=parser.get("postgres_config","port")conn=psycopg2.connect("dbname="+dbname+" user="+user+" password="+password+" host="+host,port=port)m_query="SELECT * FROM Orders;"local_filename="order_extract.csv"m_cursor=conn.cursor()m_cursor.execute(m_query)results=m_cursor.fetchall()withopen(local_filename,'w')asfp:csv_w=csv.writer(fp,delimiter='|')csv_w.writerows(results)fp.close()m_cursor.close()conn.close()# load the aws_boto_credentials valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")access_key=parser.get("aws_boto_credentials","access_key")secret_key=parser.get("aws_boto_credentials","secret_key")bucket_name=parser.get("aws_boto_credentials","bucket_name")s3=boto3.client('s3',aws_access_key_id=access_key,aws_secret_access_key=secret_key)s3_file=local_filenames3.upload_file(local_filename,bucket_name,s3_file)
Modifying the incremental version shown in the MySQL section is just as simple. All you need to do is make use of psycopg2 instead of PyMySQL.
Replicating Data Using the Write-Ahead Log
Like the MySQL binlog (as discussed in the previous section), the Postgres WAL can be used as a method of CDC for extraction. Also like the MySQL binlog, using the WAL for data ingestion in a pipeline is quite complex.
Though you can take a similar, simplified approach to the one used as an example with the MySQL binlog, I suggest using an open source distributed platform called Debezium to stream the contents of the Postgres WAL to an S3 bucket or data warehouse.
Though the specifics of configuring and running Debezium services are a topic worth dedicating an entire book to, I give an overview of Debezium and how it can be used for data ingestions in “Streaming Data Ingestions with Kafka and Debezium”. You can learn more about how it can be used for Postgres CDC there.
Extracting Data from MongoDB
This example illustrates how to extract a subset of MongoDB documents from a collection. In this sample MongoDB collection, documents represent events logged from some system such as a web server. Each document has a timestamp of when it was created, as well as a number of properties that the sample code extracts a subset of. After the extraction is complete, the data is written to a CSV file and stored in an S3 bucket so that it can be loaded into a data warehouse in a future step (see Chapter 5).
To connect to the MongoDB database, you’ll need to first install the PyMongo library. As with other Python libraries, you can install it using pip:
(env) $ pip install pymongo
You can of course modify the following sample code to connect to your own MongoDB instance and extract data from your documents. However, if you’d like to run the sample as is, you can do so by creating a MongoDB cluster for free with MongoDB Atlas. Atlas is a fully managed MongoDB service and includes a free-for-life tier with plenty of storage and computing power for learning and running samples like the one I provide. You can upgrade to a paid plan for production deployments.
You can learn how to create a free MongoDB cluster in Atlas, create a database, and configure it so that you can connect via a Python script running on your local machine by following these instructions.
You’ll need to install one more Python library named dnspython to support pymongo in connecting to your cluster hosted in MongoDB Atlas. You can install it using pip:
(env) $ pip install dnspython
Next, add a new section to the pipeline.conf file with connection information for the MongoDB instance you’ll be extracting data from. Fill in each line with your own connection details. If you’re using MongoDB Atlas and can’t recall these values from when you set up your cluster, you can learn how to find them by reading the Atlas docs.
[mongo_config] hostname = my_host.com username = mongo_user password = mongo_password database = my_database collection = my_collection
Before creating and running the extraction script, you can insert some sample data to work with. Create a file called sample_mongodb.py with the following code:
frompymongoimportMongoClientimportdatetimeimportconfigparser# load the mongo_config valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")hostname=parser.get("mongo_config","hostname")username=parser.get("mongo_config","username")password=parser.get("mongo_config","password")database_name=parser.get("mongo_config","database")collection_name=parser.get("mongo_config","collection")mongo_client=MongoClient("mongodb+srv://"+username+":"+password+"@"+hostname+"/"+database_name+"?retryWrites=true&"+"w=majority&ssl=true&"+"ssl_cert_reqs=CERT_NONE")# connect to the db where the collection residesmongo_db=mongo_client[database_name]# choose the collection to query documents frommongo_collection=mongo_db[collection_name]event_1={"event_id":1,"event_timestamp":datetime.datetime.today(),"event_name":"signup"}event_2={"event_id":2,"event_timestamp":datetime.datetime.today(),"event_name":"pageview"}event_3={"event_id":3,"event_timestamp":datetime.datetime.today(),"event_name":"login"}# insert the 3 documentsmongo_collection.insert_one(event_1)mongo_collection.insert_one(event_2)mongo_collection.insert_one(event_3)
When you execute it, the three documents will be inserted into your MongoDB collection:
(env) $ python sample_mongodb.py
Now create a new Python script called mongo_extract.py so you can add the following code blocks to it.
First, import PyMongo and Boto3 so that you can extract data from the MongoDB database and store the results in an S3 bucket. Also import the csv library so that you can structure and write out the extracted data in a flat file that’s easy to import into a data warehouse in the load step of ingestion. Finally, you’ll need some datetime functions for this example so that you can iterate through the sample event data in the MongoDB collection:
frompymongoimportMongoClientimportcsvimportboto3importdatetimefromdatetimeimporttimedeltaimportconfigparser
Next, connect to the MongoDB instance specified in the pipelines.conf file, and create a collection object where the documents you want to extract are stored:
# load the mongo_config valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")hostname=parser.get("mongo_config","hostname")username=parser.get("mongo_config","username")password=parser.get("mongo_config","password")database_name=parser.get("mongo_config","database")collection_name=parser.get("mongo_config","collection")mongo_client=MongoClient("mongodb+srv://"+username+":"+password+"@"+hostname+"/"+database_name+"?retryWrites=true&"+"w=majority&ssl=true&"+"ssl_cert_reqs=CERT_NONE")# connect to the db where the collection residesmongo_db=mongo_client[database_name]# choose the collection to query documents frommongo_collection=mongo_db[collection_name]
Now it’s time to query the documents to extract. You can do this by calling the .find() function on mongo_collection to query the documents you’re looking for. In the following example, you’ll grab all documents with a event_timestamp field value between two dates defined in the script.
Note
Extracting immutable data such as log records or generic “event” records from a data store by date range is a common use case. Although the sample code uses a datetime range defined in the script, it’s more likely you’ll pass in a datetime range to the script, or have the script query your data warehouse to get the datetime of the last event loaded, and extract subsequent records from the source data store. See “Extracting Data from a MySQL Database” for an example of doing so.
start_date=datetime.datetime.today()+timedelta(days=-1)end_date=start_date+timedelta(days=1)mongo_query={"$and":[{"event_timestamp":{"$gte":start_date}},{"event_timestamp":{"$lt":end_date}}]}event_docs=mongo_collection.find(mongo_query,batch_size=3000)
Note
The batch_size parameter in this example is set to 3000. PyMongo makes a round-trip to the MongoDB host for each batch. For example, if the result_docs Cursor has 6,000 results, it will take two trips to the MongoDB host to pull all the documents down to the machine where your Python script is running. What you set as the batch size value is up to you and will depend on the trade-off of storing more documents in memory on the system running the extract versus making lots of round trips to the MongoDB instance.
The result of the preceding code is a Cursor named event_docs that I’ll use to iterate through the resulting documents. Recall that in this simplified example, each document represents an event that was generated from a system such as a web server. An event might represent something like a user logging in, viewing a page, or submitting a feedback form. Though the documents might have dozens of fields to represent things like the browser the user logged in with, I take just a few fields for this example:
# create a blank list to store the resultsall_events=[]# iterate through the cursorfordocinevent_docs:# Include default valuesevent_id=str(doc.get("event_id",-1))event_timestamp=doc.get("event_timestamp",None)event_name=doc.get("event_name",None)# add all the event properties into a listcurrent_event=[]current_event.append(event_id)current_event.append(event_timestamp)current_event.append(event_name)# add the event to the final list of eventsall_events.append(current_event)
I’m including a default value in the doc.get() function call (–1 or None). Why? The nature of unstructured document data means that it’s possible for fields to go missing from a document altogether. In other words, you can’t assume that each of the documents you’re iterating through has an “event_name” or any other field. In those cases, tell doc.get() to return a None value instead of throwing an error.
After iterating through all the events in event_docs, the all_events list is ready to be written to a CSV file. To do so, you’ll make use of the csv module, which is included in the standard Python distribution and was imported earlier in this example:
export_file="export_file.csv"withopen(export_file,'w')asfp:csvw=csv.writer(fp,delimiter='|')csvw.writerows(all_events)fp.close()
Now, upload the CSV file to the S3 bucket that you configured in “Setting Up Cloud File Storage”. To do so, use the Boto3 library:
# load the aws_boto_credentials valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")access_key=parser.get("aws_boto_credentials","access_key")secret_key=parser.get("aws_boto_credentials","secret_key")bucket_name=parser.get("aws_boto_credentials","bucket_name")s3=boto3.client('s3',aws_access_key_id=access_key,aws_secret_access_key=secret_key)s3_file=export_files3.upload_file(export_file,bucket_name,s3_file)
That’s it! The data you extracted from the MongoDB collection is now sitting in the S3 bucket waiting to be loaded into the data warehouse or other data store. If you used the sample data provided, the contents of export_file.csv will look something like this:
1|2020-12-13 11:01:37.942000|signup 2|2020-12-13 11:01:37.942000|pageview 3|2020-12-13 11:01:37.942000|login
See Chapter 5 for more on loading the data into the data store of your choice.
Extracting Data from a REST API
REST APIs are a common source to extract data from. You may need to ingest data from an API that your organization created and maintains, or from an API from an external service or vendor that your organization uses, such as Salesforce, HubSpot, or Twitter. No matter the API, there’s a common pattern for data extraction that I’ll use in the simple example that follows:
-
Accept the response, which is most likely formatted in JSON.
-
Parse the response and “flatten” it into a CSV file that you can later load into the data warehouse.
Note
Though I am parsing the JSON response and storing it in a flat file (CSV), you can also save the data in JSON format for loading into your data warehouse. For the sake of simplicity, I’m sticking to the pattern of this chapter and using CSV files. Please consult Chapter 5 or your data warehouse documentation for more on loading data in a format other than CSV.
In this example, I’ll connect to an API called Open Notify. The API has several endpoints, each returning data from NASA about things happening in space. I’ll query the endpoint that returns the next five times that the International Space Station (ISS) will pass over the given location on Earth.
Before I share the Python code for querying the endpoint, you can see what the output of a simple query looks like by typing the following URL into your browser:
http://api.open-notify.org/iss-pass.json?lat=42.36&lon=71.05
The resulting JSON looks like this:
{
"message": "success",
"request": {
"altitude": 100,
"datetime": 1596384217,
"latitude": 42.36,
"longitude": 71.05,
"passes": 5
},
"response": [
{
"duration": 623,
"risetime": 1596384449
},
{
"duration": 169,
"risetime": 1596390428
},
{
"duration": 482,
"risetime": 1596438949
},
{
"duration": 652,
"risetime": 1596444637
},
{
"duration": 624,
"risetime": 1596450474
}
]
}
The goal of this extraction is to retrieve the data in the response and format it in a CSV file with one line for each time and duration of each pass that the ISS will make over the lat/long pair. For example, the first two lines of the CSV file will be as follows:
42.36,|71.05|623|1596384449 42.36,|71.05|169|1596390428
To query the API and handle the response in Python, you’ll need to install the requests library. requests makes HTTP requests and responses easy to work with in Python. You can install it with pip:
(env) $ pip install requests
Now, you can use requests to query the API endpoint, get back the response, and print out the resulting JSON, which will look like what you saw in your browser:
importrequestslat=42.36lon=71.05lat_log_params={"lat":lat,"lon":lon}api_response=requests.get("http://api.open-notify.org/iss-pass.json",params=lat_log_params)(api_response.content)
Instead of printing out the JSON, I’ll iterate through the response, parse out the values for duration and risetime, write the results to a CSV file, and upload the file to the S3 bucket.
To parse the JSON response, I’ll import the Python json library. There’s no need to install it as it comes with the standard Python installation. Next, I’ll import the csv library, which is also included in the standard Python distribution for writing the CSV file. Finally, I’ll use the configparser library to get the credentials required by the Boto3 library to upload the CSV file to the S3 bucket:
importrequestsimportjsonimportconfigparserimportcsvimportboto3
Next, query the API just as you did before:
lat=42.36lon=71.05lat_log_params={"lat":lat,"lon":lon}api_response=requests.get("http://api.open-notify.org/iss-pass.json",params=lat_log_params)
Now, it’s time to iterate through the response, store the results in a Python list called all_passes, and save the results to a CSV file. Note that I also store the lat and long from the request even though they are not included in the response. They are needed on each line of the CSV file so that the pass times are associated with the correct lat and long when loaded into the data warehouse:
# create a json object from the response contentresponse_json=json.loads(api_response.content)all_passes=[]forresponseinresponse_json['response']:current_pass=[]#store the lat/log from the requestcurrent_pass.append(lat)current_pass.append(lon)# store the duration and risetime of the passcurrent_pass.append(response['duration'])current_pass.append(response['risetime'])all_passes.append(current_pass)export_file="export_file.csv"withopen(export_file,'w')asfp:csvw=csv.writer(fp,delimiter='|')csvw.writerows(all_passes)fp.close()
Finally, upload the CSV file to the S3 bucket using the Boto3 library:
# load the aws_boto_credentials valuesparser=configparser.ConfigParser()parser.read("pipeline.conf")access_key=parser.get("aws_boto_credentials","access_key")secret_key=parser.get("aws_boto_credentials","secret_key")bucket_name=parser.get("aws_boto_credentials","bucket_name")s3=boto3.client('s3',aws_access_key_id=access_key,aws_secret_access_key=secret_key)s3.upload_file(export_file,bucket_name,export_file)
Streaming Data Ingestions with Kafka and Debezium
When it comes to ingesting data from a CDC system such as MySQL binlogs or Postgres WALs, there’s no simple solution without some help from a great framework.
Debezium is a distributed system made up of several open source services that capture row-level changes from common CDC systems and then streams them as events that are consumable by other systems. There are three primary components of a Debezium installation:
-
Apache Zookeeper manages the distributed environment and handles configuration across each service.
-
Apache Kafka is a distributed streaming platform that is commonly used to build highly scalable data pipelines.
-
Apache Kafka Connect is a tool to connect Kafka with other systems so that the data can be easily streamed via Kafka. Connectors are built for systems like MySQL and Postgres and turn data from their CDC systems (binlogs and WAL) into Kakfa topics.
Kafka exchanges messages that are organized by topic. One system might publish to a topic, while one or more might consume, or subscribe to, the topic.
Debezium ties these systems together and includes connectors for common CDC implementations. For example, I discussed the challenges for CDC in “Extracting Data from a MySQL Database” and “Extracting Data from a PostgreSQL Database”. Thankfully, there are connectors already built to “listen” to the MySQL binlog and Postgres WAL. The data is then routed through Kakfa as records in a topic and consumed into a destination such as an S3 bucket, Snowflake, or Redshift data warehouse using another connector. Figure 4-1 illustrates an example of using Debezium, and its individual components, to send the events created by a MySQL binlog into a Snowflake data warehouse.
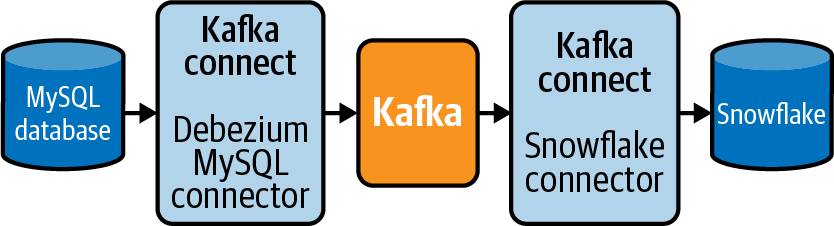
Figure 4-1. Using components of Debezium for CDC from MySQL to Snowflake.
As of this writing, there are a number of Debezium connectors already built for source systems that you may find yourself needing to ingest from:
-
MongoDB
-
MySQL
-
PostgreSQL
-
Microsoft SQL Server
-
Oracle
-
Db2
-
Cassandra
There are also Kafka Connect connectors for the most common data warehouses and storage systems, such as S3 and Snowflake.
Though Debezium, and Kafka itself, is a subject that justifies its own book, I do want to point out its value if you decide that CDC is a method you want to use for data ingestion. The simple example I used in the MySQL extraction section of this chapter is functional; however, if you want to use CDC at scale, I highly suggest using something like Debezium rather than building an existing platform like Debezium on your own!
Tip
The Debezium documentation is excellent and a great starting point for learning about the system.
Get Data Pipelines Pocket Reference now with the O’Reilly learning platform.
O’Reilly members experience books, live events, courses curated by job role, and more from O’Reilly and nearly 200 top publishers.

