The community lab: A student’s door to opportunity
Shreya Thiagarajan describes the invaluable experience she gained while working in a local community lab.
 Microscope. (source: Pixabay)
Microscope. (source: Pixabay)
In what seems like a revolutionary rise in citizen science, hackerspaces (fondly called
community labs by their users) have emerged at the forefront as a potential solution to the lack
of scientific resources available to ordinary citizens. They provide everyone with access to
ample resources and materials needed to kickstart a project that may otherwise require years of
funding to begin. Hackerspaces are on the rise nationwide, popping up in several big cities all
over the United States such as New York, Los Angeles, and San Francisco.
Hackerspaces vary in nearly every aspect, from size to accessibility to funding. Many
hackerspaces specialize in a particular field of science, such as nanotechnology or
computational biology, while many others are hosts to projects in a variety of different scientific
subfields. The consensus seems to be that the typical hackerspace is a location for a meeting of
the minds, that is, an open resource center for a scientific community to meet and exchange
ideas about potential laboratory-common projects. For example, Counter Culture labs, a
hackerspace in Berkeley, California, plays host to the Open Insulin project, one that the most
lab members work on as a laboratory-common project. In most cases, hackerspaces specialize
in one or two projects the entire community works together on like the Open Insulin project, but
they usually also serve as laboratories for their users to conduct individual projects in.
Such individual projects have bloomed into successful products, some of which can even
be found on the market today. What most citizen scientists have not considered, however, is the
remarkable impact these hackerspaces have on college students and high-school students like
me.
One such hackerspace is BioCurious, located in Sunnyvale, California, deep in the heart
of the Silicon Valley. Not only does BioCurious hold regular classes to teach students basic
laboratory procedures and techniques, but it also encourages them to take part in its various
community projects, many of which are submitted to the annual International Genetically
Engineered Machine (iGEM) jamboree, a contest that attracts teams from universities and
hackerspaces worldwide to present their innovative ideas at a conference-style event in Boston.
The iGEM jamboree is unique because it fosters collaboration among participating teams by
allowing them to submit their engineered biological components to their database from which
future teams can request certain “bio-blocks” to use in their projects. In 2014, when iGEM had
just begun accepting community labs as participants, BioCurious won a gold medal in its
category for its Real Vegan Cheese project. The following year, it was able to secure a bronze
medal in its category for its Bio Sunblock project.
Having joined BioCurious in the December of 2014 I was unfortunately unable to
participate in the Real Vegan Cheese project. I was, however, graciously invited to participate in
the 2015 iGEM project, first helping to compile a list of possible topics to do further research on.
It was here that my involvement in BioCurious’s community projects and my interest in citizen
science began.
My first dosage of scholarly research came from perusing various publications and
journal articles to gain information about the topic I had chosen to vouch for from an ideas list
that included everything from inducing bacteria to grow into three-dimensional shapes to using natural compounds as biological ink. Along with my high-school and college peers, I was able to
design a poster-style slide presentation detailing specific information about each project topic,
including the required materials, feasibility, and societal impact. When the voting was
completed, only one project remained.
Our idea of producing biological sunblock-like compounds stemmed from the issue of
chemical pollution in freshwater rivers and lakes, as well as one of the most important
ecosystems on our planet, the ocean. Information from a study published by the journal
Ecotoxicology and reported by the National Ocean Service indicated that certain compounds in
sunscreen that washed off of swimmers in the ocean were contributing to coral bleaching and
reef death. Our goal was to find a method to keep ourselves protected from dangerous
ultraviolet rays while simultaneously combating the effects of chemical pollution in the ocean.
Our team noted that several marine organisms such as algae and some coral species
already produced UV-protectant substances to shield growing polyps from harmful solar
radiation. Mycosporine-like amino acids (MAAs), as they were called, could be induced upon
prolonged exposure to ultraviolet light and were full of anti-oxidative compounds that fought
radiation-caused skin damage.
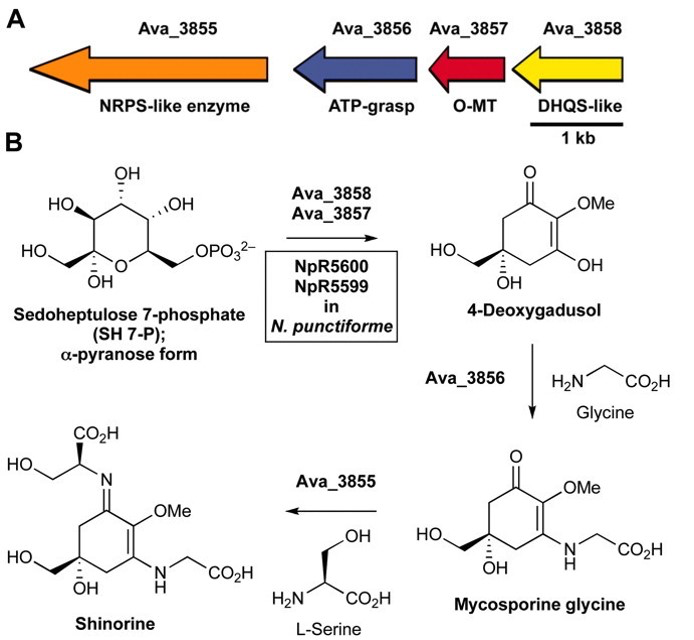
Shinorine, a certain MAA produced by the cyanobacteria Anabaena variabilis , had
already been studied extensively and had shown promising results as a potential biological
sunblock. The gene for the compound in A. variabilis was first discovered in 2010 by
researchers Emily Balskus and Christopher Walsh at Harvard University; and in 2012, the
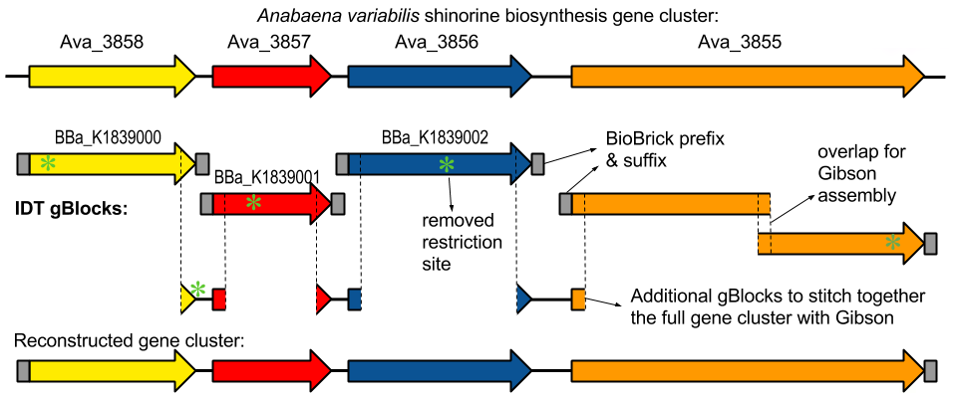
University of Minnesota iGEM team succeeded in creating the gene cluster containing the
shinorine producing gene (Figure 1-1 and Figure 1-2). Synthetic shinorine production in Escherichia coli
containing the transformed gene cluster was yet to be observed, so our team took to the task.
Up to this point, the most cutting-edge scientific procedures I had ever done were
preparing agar plates and culturing bacteria. I was understandably surprised then when I (along
with some responsible adults, of course) was assigned, as a learning opportunity, the task of
transforming the completed gene cluster into E. coli, using the “bio-blocks,” or editing tools,
provided by IDT, an iGEM sponsor. Gene editing had only been briefly mentioned in my
freshman biology class, so all this time I had thought of the process as an expensive and
extremely delicate procedure only performed in the most secure and highly funded of
laboratories. Yet I was handed a series of different-sized micropipettes and small vials of liquid
presumably holding the gene cluster that needed to be transformed into the bacteria.
Not many high-school students can boast about having had the opportunity to edit
genes, and I owed my privilege to BioCurious for placing such resources into my hands.
Although accuracy in results was what we aimed for, the team emphasized education as a
critical component of the experimental process. It was under their guidance that one Saturday
morning, I was able to transform the shinorine-producing gene into a culture of E. coli with little
difficulty.
Over the following six months before the jamboree, I could be found at BioCurious every
Saturday morning helping perform spot assays (a process that uses serial dilution of a bacterial
culture sample to measure bacterial growth) in the fume hood at the corner of the lab or fiddling
with the DeNovix spectrophotometer.
Two years after the project won BioCurious a bronze medal at the 2015 iGEM jamboree,
the project was discontinued, yet it continues to have an impact on my life and my opportunities
as a student researcher. My work at BioCurious has opened several doors for me, and not only
has my experience helped me obtain research positions in university laboratories, but with
BioCurious’s help, I was able to carry out two projects of my own, both of which I submitted to
the local Synopsis science fair.
My first project focused on the neurodegenerative disease amyotrophic lateral sclerosis
(ALS), whose growing prevalence in the elderly population, combined with the popularity of the
ALS ice bucket challenge at the time, piqued my interest around the same time the deadline for
submissions to the local science fair was approaching. The rapid progression of the disease is closely correlated to an increase in the production of the protein TDP-43. However, it was the
clumping of this protein in motor neurons that is believed to lead to rapid cell death and
paralysis (Mackenzie, Ian R.A.; Rademakers, Rosa).
From reading journal articles, I discovered that a particular natural compound known as
curcumin had been effective in inhibiting the clumping of proteins in motor neurons that lead to
similar diseases such as Parkinson’s disease and Huntington’s disease. In fact, curcumin was
proven by a research team in UCLA and confirmed by two other scientists to have disbanded
beta-amyloid plaque buildup in cell models with Alzheimer’s disease (Yang, Fusheng, et al.,
2004) With this information in mind, I decided to test the effects of curcumin on the clumping of
TDP-43 in ALS yeast models.
This particular project was especially unforgettable since it was the first time I would
complete a science project that I myself designed and carried out on my own. Of course, I could
not have accomplished a single part of the project without the resources and materials provided
by BioCurious which were so generously shared by their respective owners for the whole lab to
use. Additionally since it was my first time performing some other laboratory procedures (using
the fluorescent microscope, gel electrophoresis, etc.) I had significant help from several other
lab members (mentioned in the acknowledgments) to carry out these procedures. In time, these
resources allowed me to finish my project before the regional science fair, and with the help I
received from my fellow lab members, I was able to present my data seamlessly to all who
chose to listen.
The following year, another pressing issue took root in my mind, prompting me to
research its implications and impact on society today: antibiotic resistance, particularly the
presence of antibiotics in livestock feed. It was a “solution” that had been devised to combat the
growing prevalence of disease in livestock that resulted from crowded and dilapidated pens and
uncleaned feed. Through the repeated administration of antibiotics to livestock through crops
and feed, mutant bacteria with innate resistance have survived with each increased
concentration of antibiotics and found their way into the animals we interact with and the meat
we consume.
Since traditional antibiotics were the problem itself, other chemicals proven to be
antimicrobial would not serve as an adequate solution since bacteria could evolve a resistance
to those chemicals as well. An ideal solution would be one that evolved along with the
resistance in bacteria. That ideal solution was the bacteriocin, a category of compounds
naturally produced by some bacteria species to kill other bacterial species that compete for the
same resources. Since these bacteria coexist, a developing resistance in one bacteria species
could trigger evolution of the other bacteria species, favoring individuals that produce an
“evolved” bacteriocin that could now kill the resistant bacteria.
One species of bacteria Lactococcus lactis, commonly found in the cow’s digestive tract,
produces a specific bacteriocin called nisin. Past studies proved nisin as a bacteria effective in
killing gram-positive bacteria, including two particular species that are commonly found in animal
feed.
Though nisin itself would have been an optimal choice as a deterrent to use in my
experiment, past studies had proven its effectiveness in reducing the growth of bacteria. Thus, I
decided I would primarily research the effects of nisin combined with either ascorbic acid or acetic acid on bacterial growth. Acetic acid, or vinegar, was itself antimicrobial, so I hoped a
combination of the two would enhance nisin’s performance.
The results from my research were enough to win me a special award at the regional
science fair and a first-place cash award from the Northern California Institute of Food
Technology. I give credit for both to BioCurious for providing me with the materials and help I
needed to succeed in my scientific endeavors.
Overall, my past two years at Biocurious have opened numerous doors. I was able to
present at the annual Bay Area Makerfaire on behalf of the laboratory. Maker Faire is one of the
area’s most popular annual attractions and plays host to hundreds of booths and organizations
that flock to the event to display their creativity in various ways. I still remember my excitement
from being given a free ticket to the event, a perk that all participating organizations received for
sharing their innovations with the public. What stuck in my mind as one of the most rewarding
moments of the experience was watching two children, still in elementary school, waddle up to
our booth with excitement. Their eyes widened in fascination as I spoke to them about how we
had stripped a sheep heart of cells, leaving a protein scaffold behind that could be used as a
template to create a functional human heart out of stem cells.
Their mother approached me and began inquiring about the lab as her children worked
on extracting their own DNA from their cheek cells, an activity we had designed for interested
children and adults alike to try. I recall being congratulated and commended for my passion for
science and for my will to take initiative and accomplish so much at a rather young age.
The truth is most high-school students are interested in cutting-edge research, and those
who are currently uninterested are soon to change when exposed to the kind of technology that
is changing the way we innovate. That is where community labs make their biggest impact,
inviting anyone, regardless of his or her experience and interests, to make dreams come to life.
I think I can speak for all my high-school peers at Biocurious that community labs have been a
key part of our development as future scientists.
I would like to thank Maria Chavez for letting me join BioCurious as a high-school
student and participate in the 2015 iGEM project, Patrick D’haeseleer and Jay Hanson for
guiding me through each step of the experimental process in the Bio Sunblock project, and
Johan Sosa for assisting me in both my personal projects and allowing me to help him on the
Real Vegan Cheese project. Special thanks to Eric Harness for teaching me how to use the
autoclave and the fluorescent microscope.
